Deposition Date
2021-02-13
Release Date
2021-12-01
Last Version Date
2024-11-06
Entry Detail
PDB ID:
7NIS
Keywords:
Title:
1918 H1N1 Viral influenza polymerase heterotrimer with Nb8192 core
Biological Source:
Source Organism(s):
Camelidae mixed library (Taxon ID: 1579311)
Influenza A virus (strain A/Brevig Mission/1/1918 H1N1) (Taxon ID: 88776)
Staphylococcus aureus (Taxon ID: 1280)
Influenza A virus (strain A/Brevig Mission/1/1918 H1N1) (Taxon ID: 88776)
Staphylococcus aureus (Taxon ID: 1280)
Expression System(s):
Method Details:
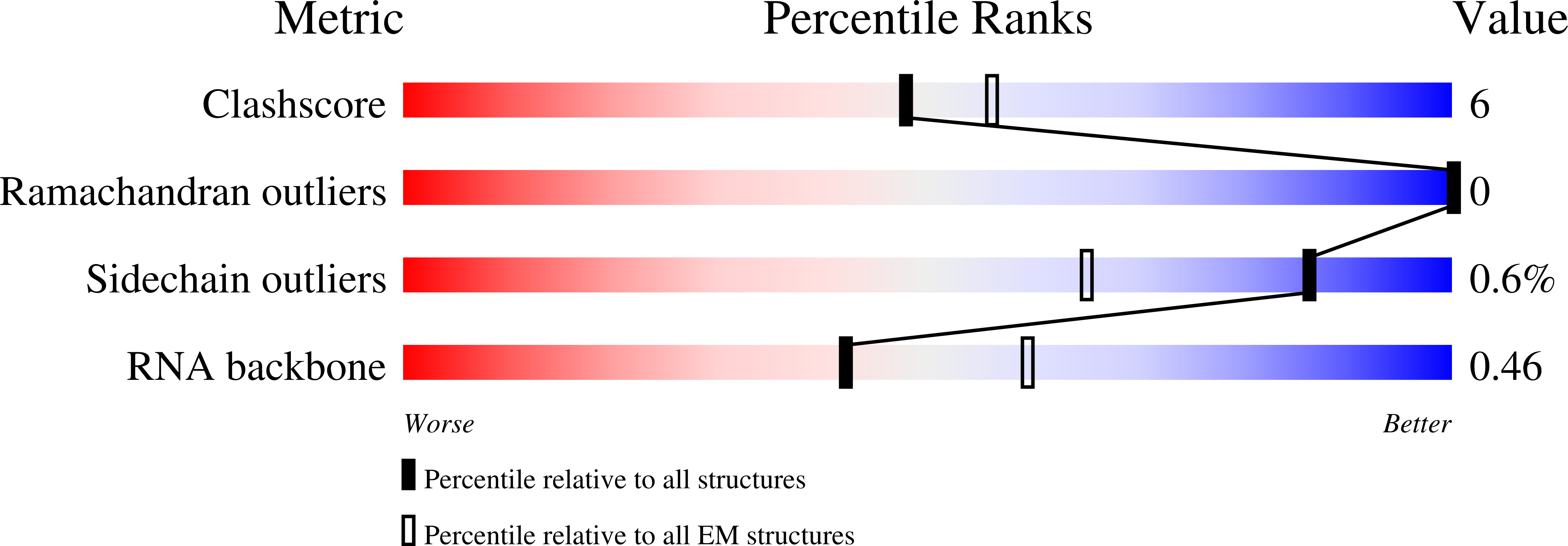
Experimental Method:
Resolution:
5.96 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE