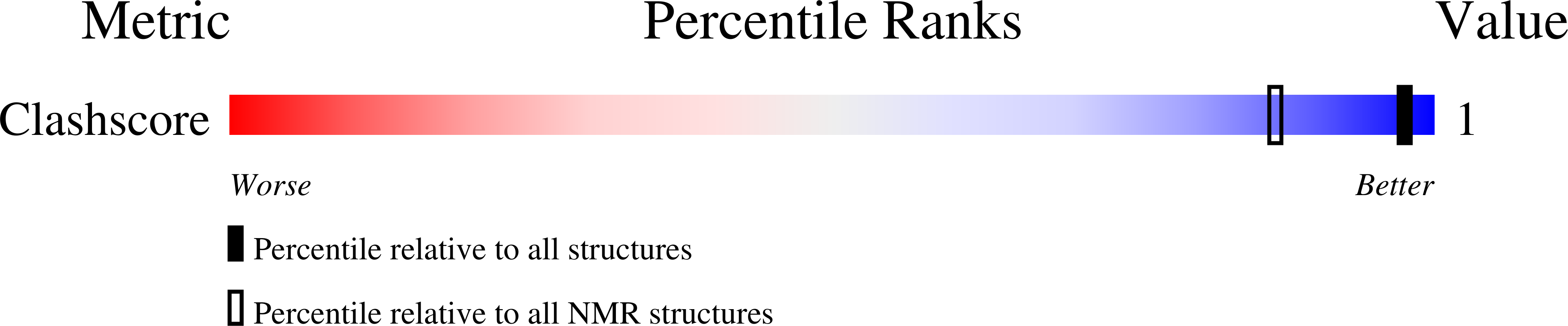
Deposition Date
2021-01-27
Release Date
2022-03-02
Last Version Date
2024-06-19
Entry Detail
PDB ID:
7NBP
Keywords:
Title:
Solution structure of DNA duplex containing a 7,8-dihydro-8-oxo-1,N6-ethenoadenine base modification that induces exclusively A->T transversions in Escherichia coli
Biological Source:
Source Organism(s):
synthetic construct (Taxon ID: 32630)
Method Details:
Experimental Method:
Conformers Calculated:
10
Conformers Submitted:
10
Selection Criteria:
all calculated structures submitted