Deposition Date
2021-05-03
Release Date
2021-07-14
Last Version Date
2024-05-29
Entry Detail
PDB ID:
7MOQ
Keywords:
Title:
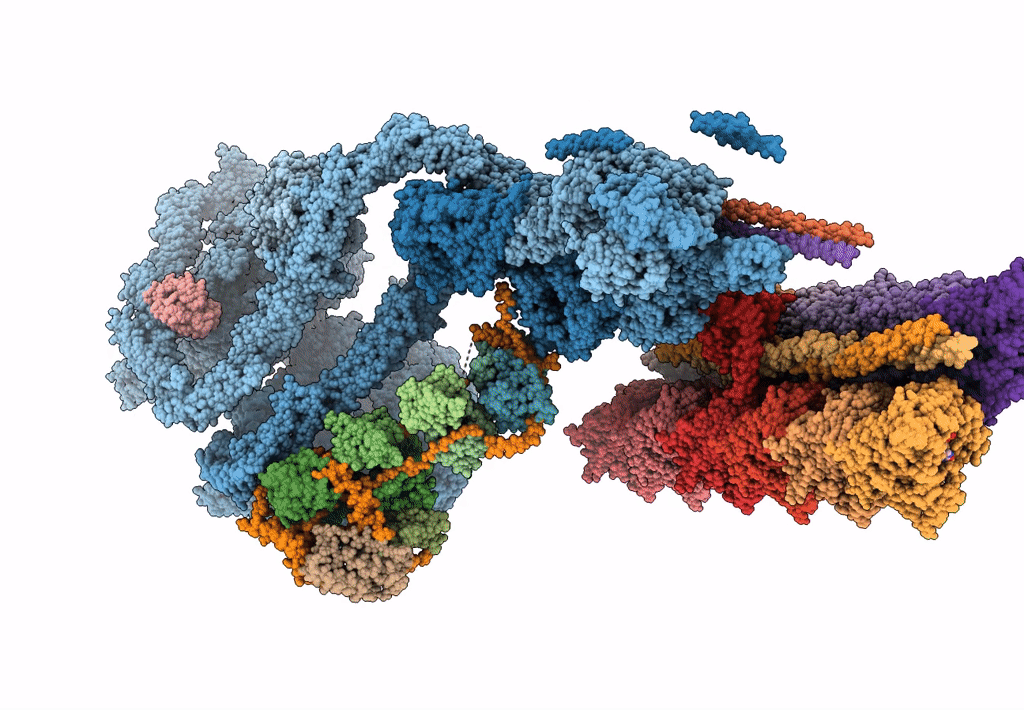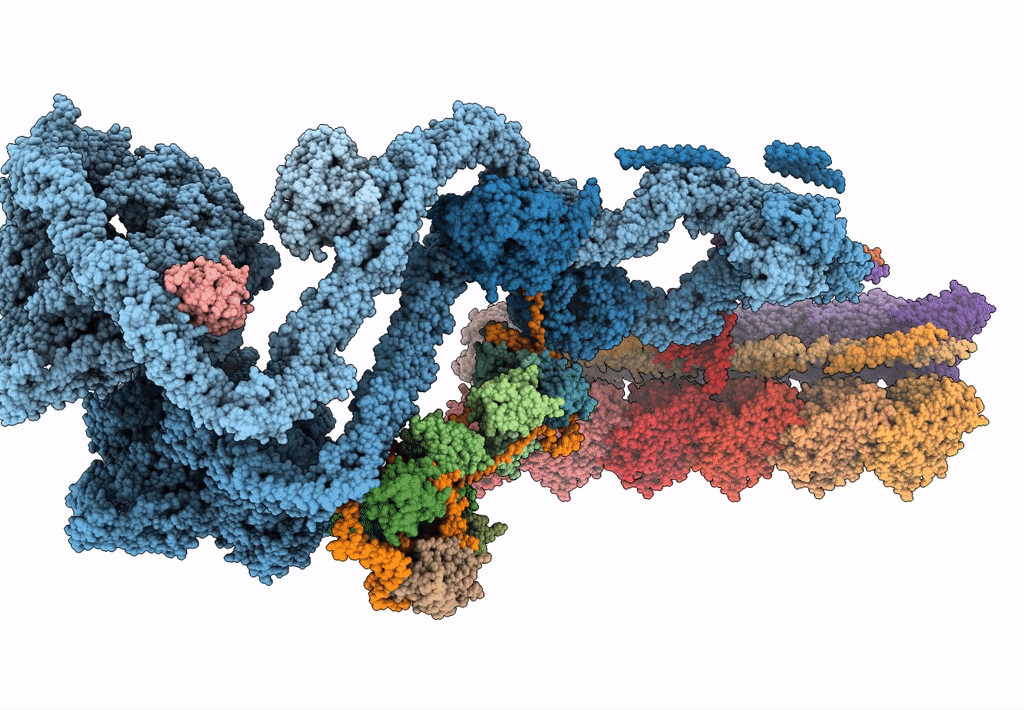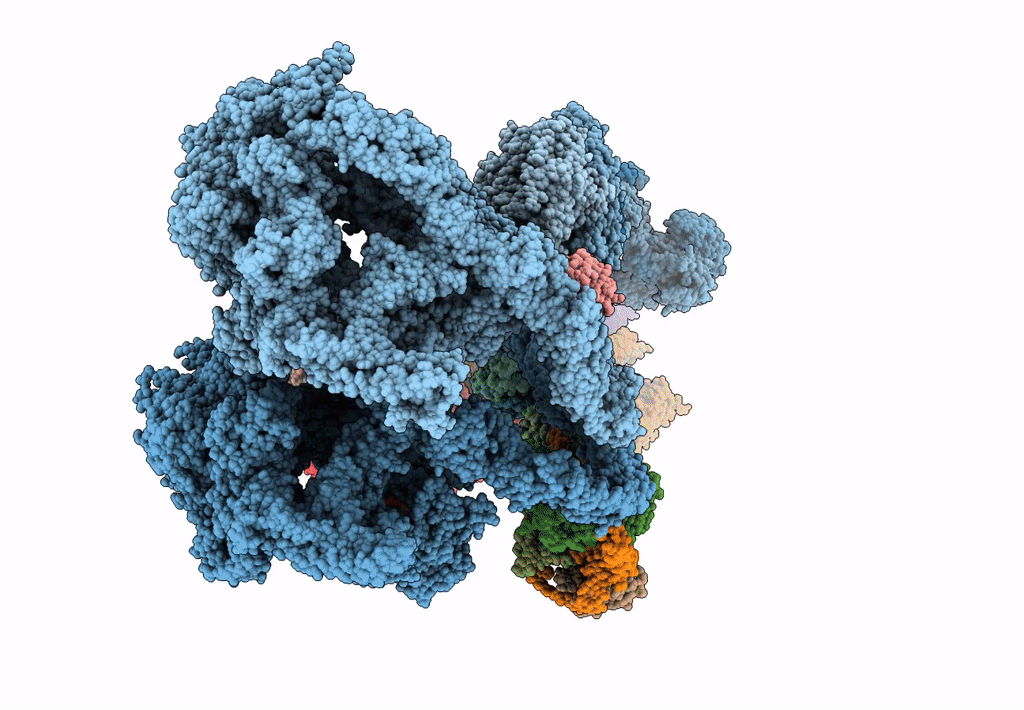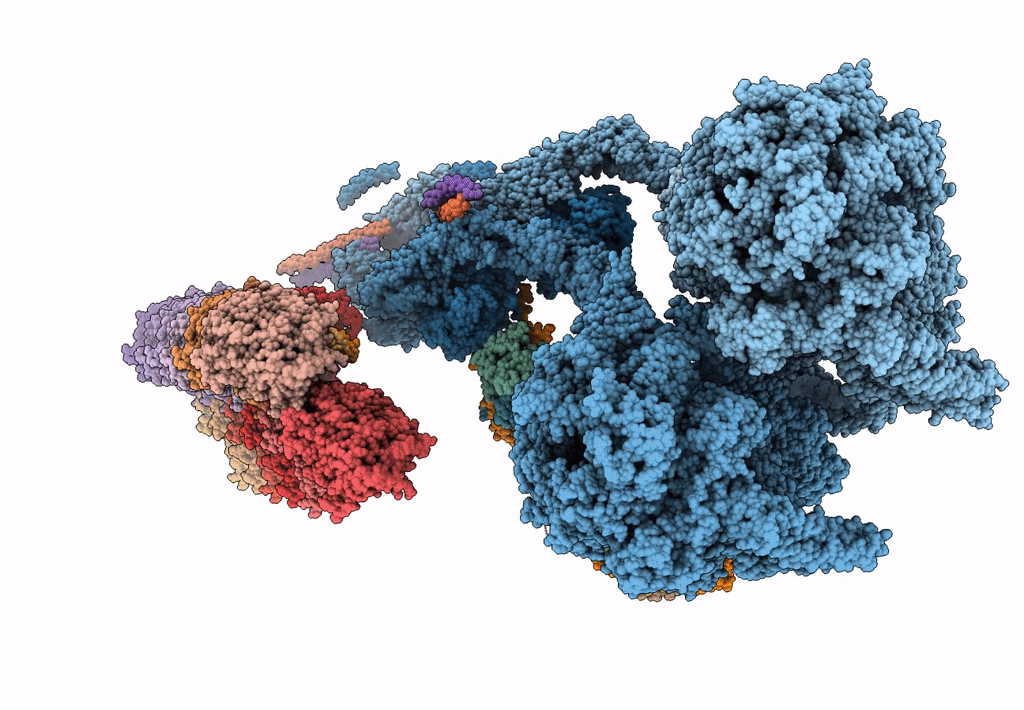
The structure of the Tetrahymena thermophila outer dynein arm on doublet microtubule
Biological Source:
Source Organism:
Tetrahymena thermophila CU428 (Taxon ID: 452467)
Method Details:
Experimental Method:
Resolution:
8.00 Å
Aggregation State:
FILAMENT
Reconstruction Method:
SINGLE PARTICLE


