Deposition Date
2021-04-16
Release Date
2021-05-26
Last Version Date
2024-05-29
Entry Detail
PDB ID:
7MI3
Keywords:
Title:
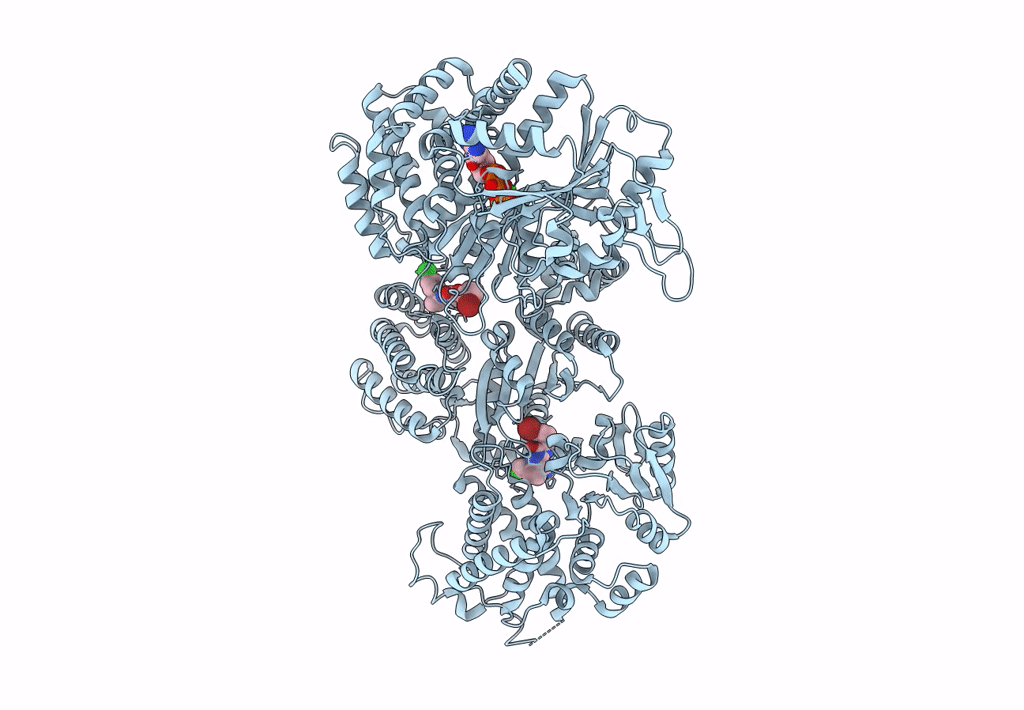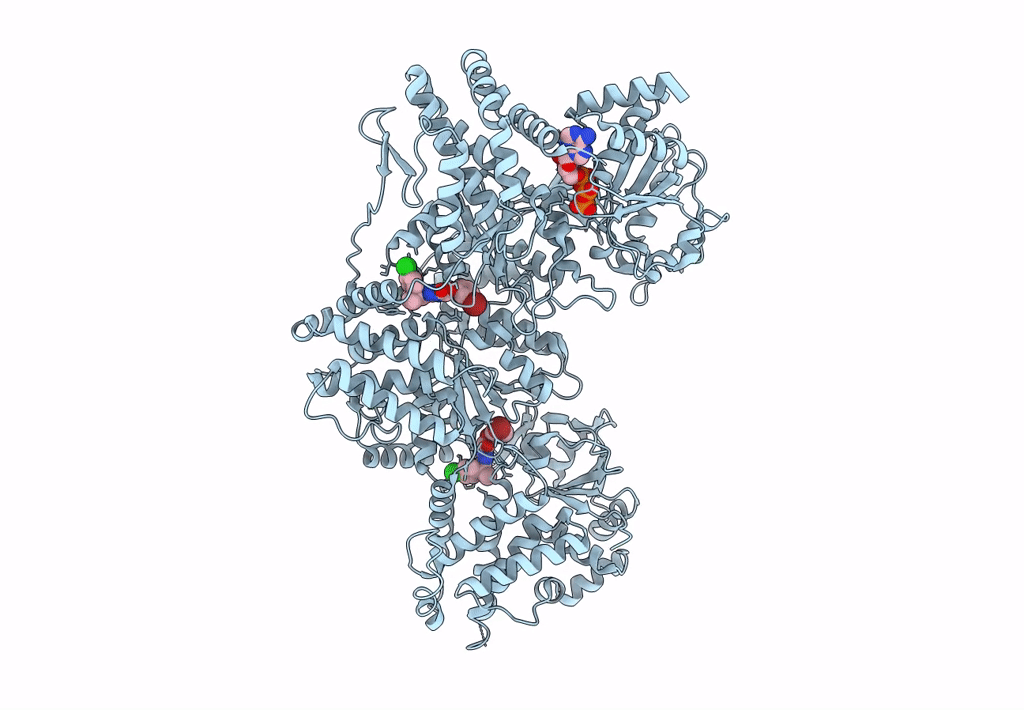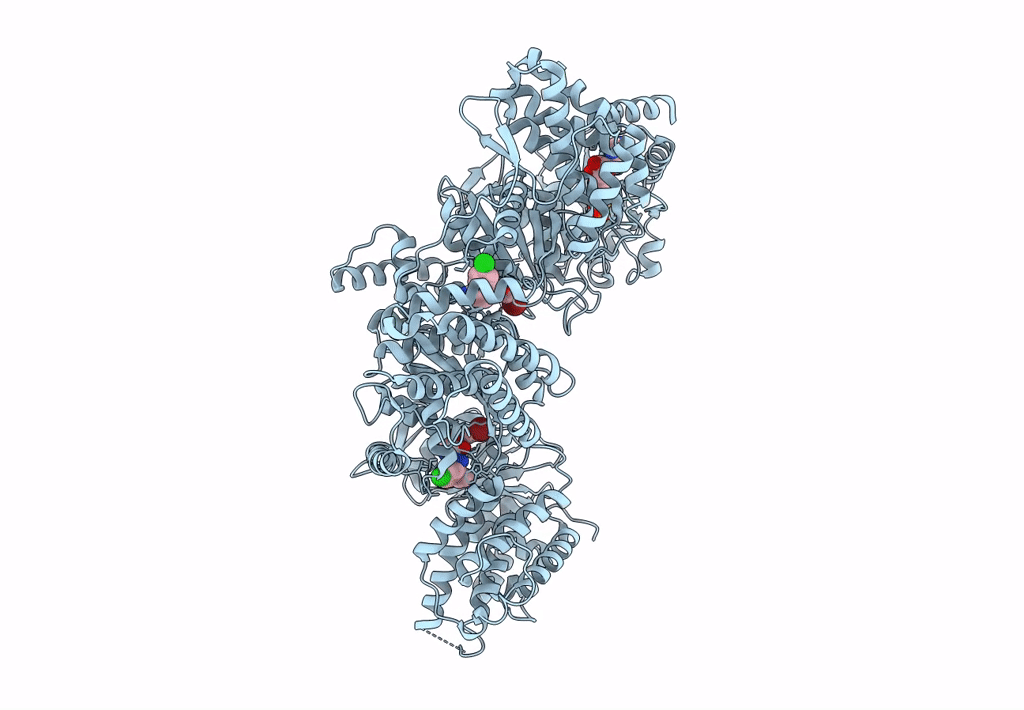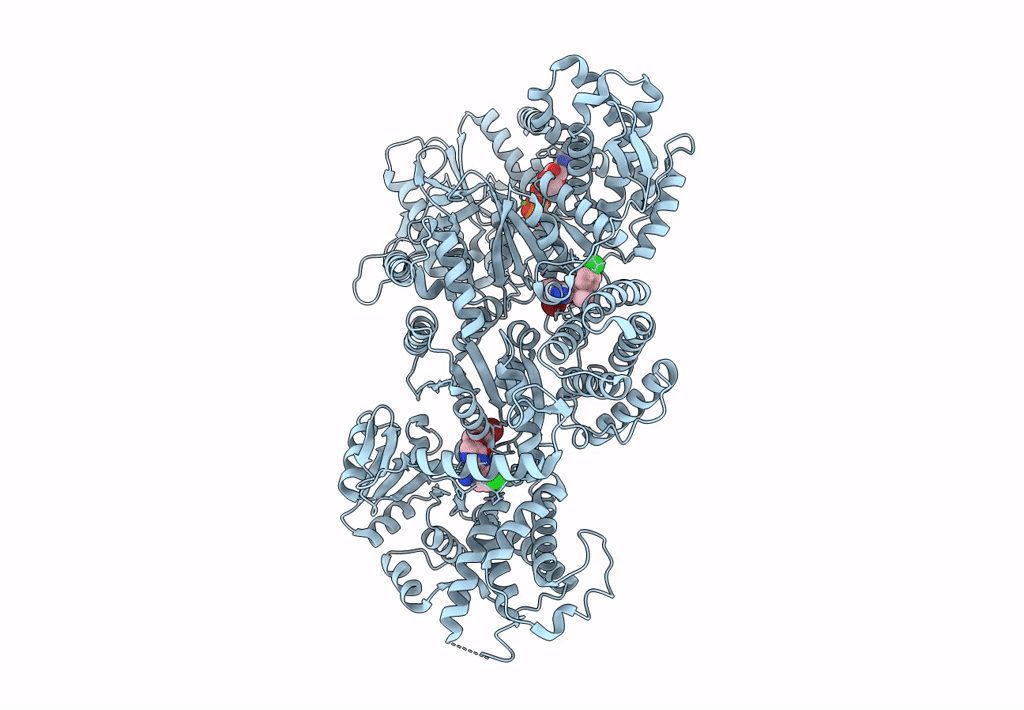
Signal subtracted reconstruction of AAA2, AAA3, and AAA4 domains of dynein in the presence of a pyrazolo-pyrimidinone-based compound, Model 4
Biological Source:
Source Organism(s):
Saccharomyces cerevisiae (Taxon ID: 4932)
Enterobacteria phage T4 (Taxon ID: 10665)
Enterobacteria phage T4 (Taxon ID: 10665)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.50 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


