Deposition Date
2021-03-26
Release Date
2021-11-24
Last Version Date
2024-11-13
Entry Detail
PDB ID:
7M76
Keywords:
Title:
Room Temperature XFEL Crystallography reveals asymmetry in the vicinity of the two phylloquinones in Photosystem I
Biological Source:
Source Organism(s):
Thermosynechococcus elongatus (strain BP-1) (Taxon ID: 197221)
Method Details:
Experimental Method:
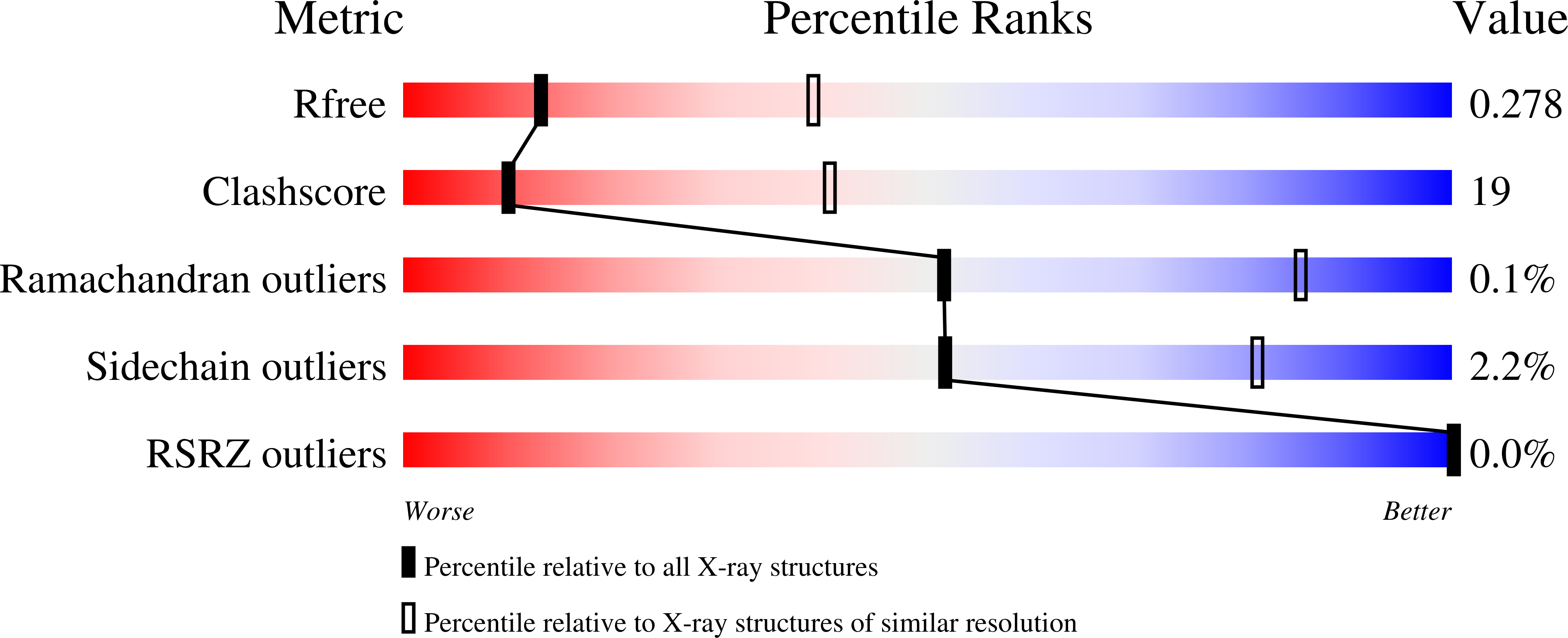
Resolution:
3.00 Å
R-Value Free:
0.27
R-Value Work:
0.26
R-Value Observed:
0.26
Space Group:
P 63