Deposition Date
2021-03-19
Release Date
2021-12-15
Last Version Date
2023-10-18
Entry Detail
PDB ID:
7M3T
Keywords:
Title:
Crystallographic structure of a cubic crystal of STMV (80.7 degree rotation about 111) grown from chloride
Biological Source:
Source Organism(s):
Satellite tobacco mosaic virus (Taxon ID: 12881)
Method Details:
Experimental Method:
Resolution:
3.20 Å
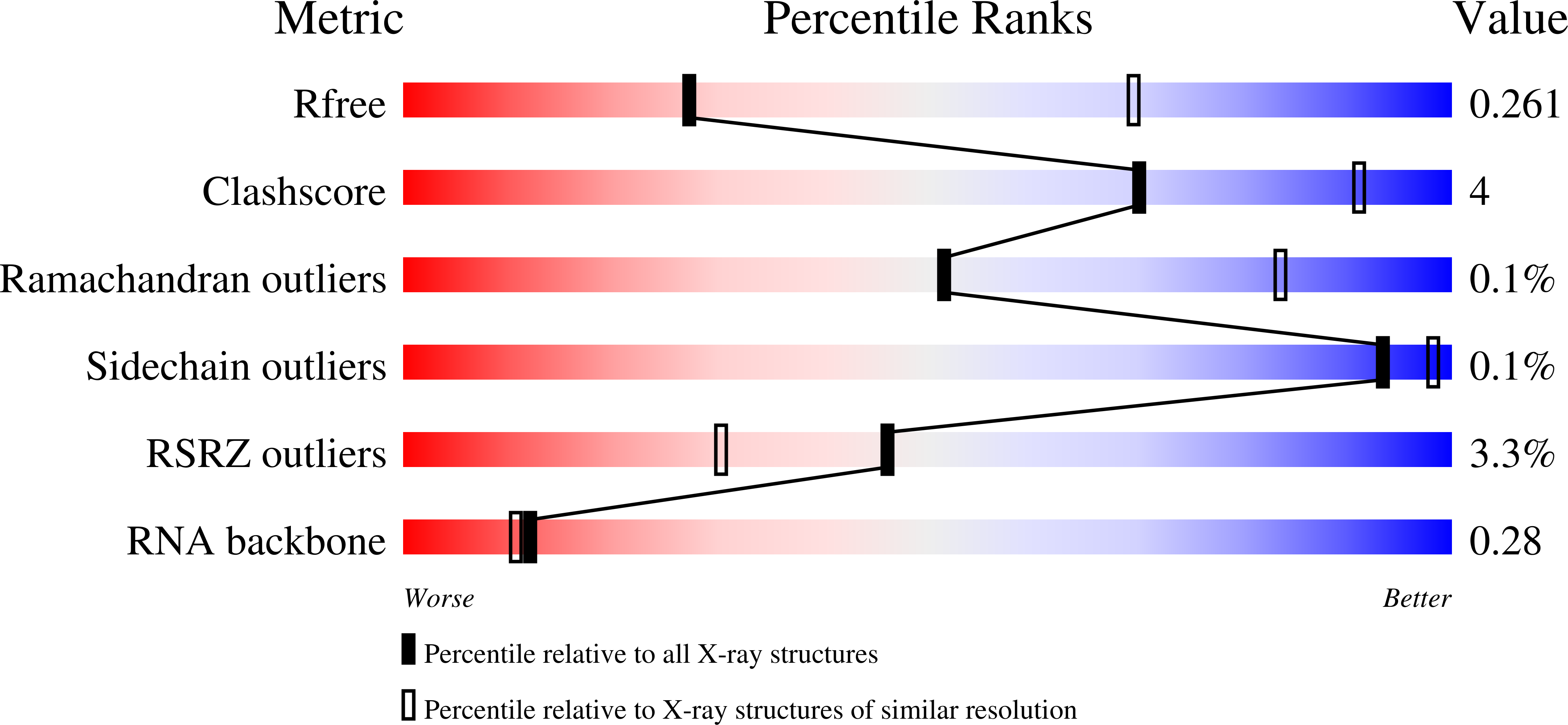
R-Value Free:
0.25
R-Value Work:
0.21
R-Value Observed:
0.21
Space Group:
P 2 3