Deposition Date
2020-12-30
Release Date
2021-03-03
Last Version Date
2023-10-18
Entry Detail
PDB ID:
7L7Z
Keywords:
Title:
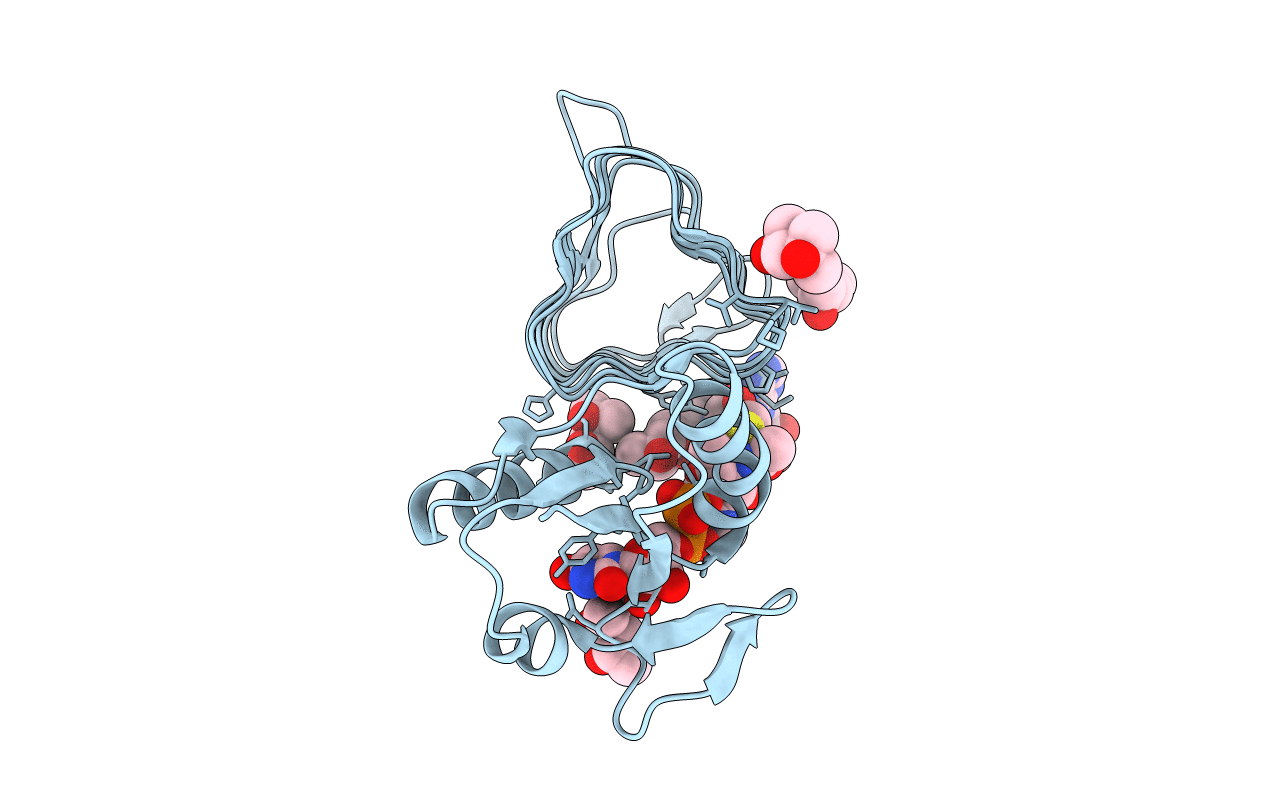
x-ray structure of the N-acetyltransferase Pcryo_0637 from psychrobacter cryohalolentis in the presence of coenzyme A and UDP-di-N-acetyl-bacillosamine
Biological Source:
Source Organism(s):
Expression System(s):
Method Details:
Experimental Method:
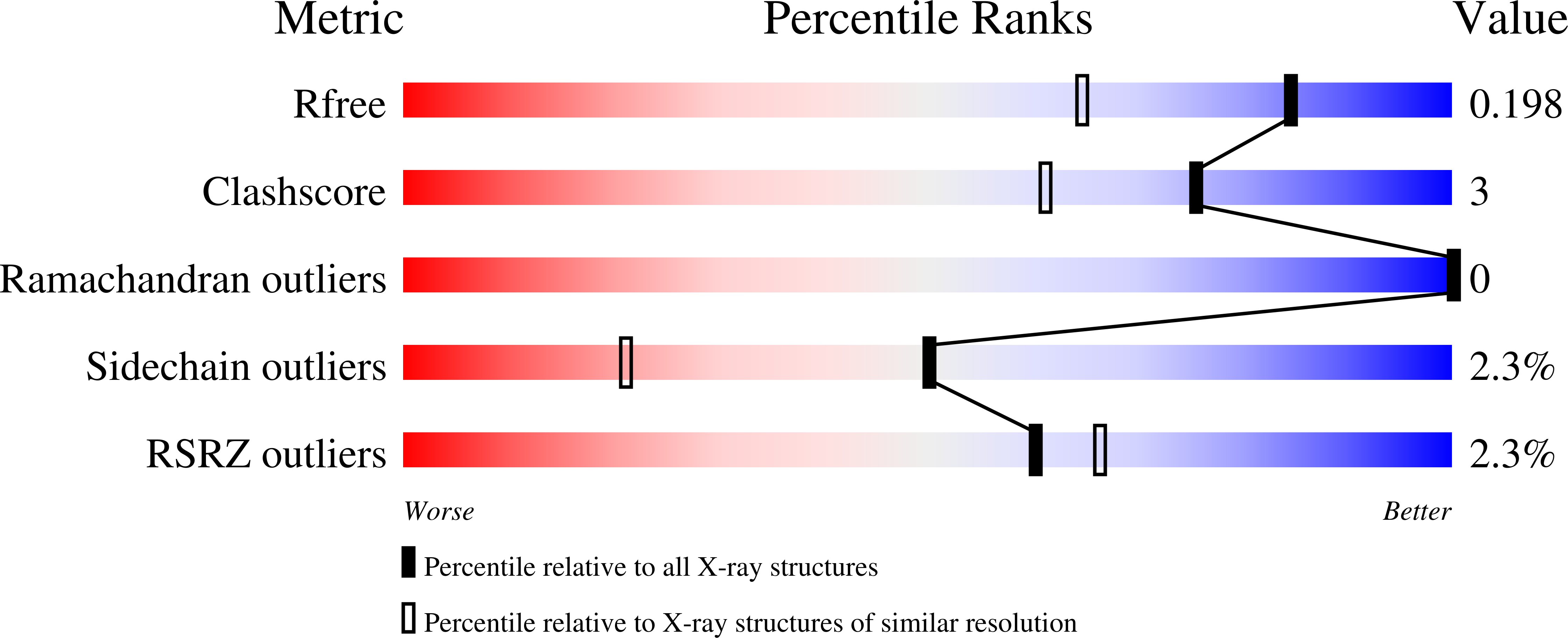
Resolution:
1.55 Å
R-Value Free:
0.19
R-Value Work:
0.16
Space Group:
H 3