Abstact
UNLABELLED
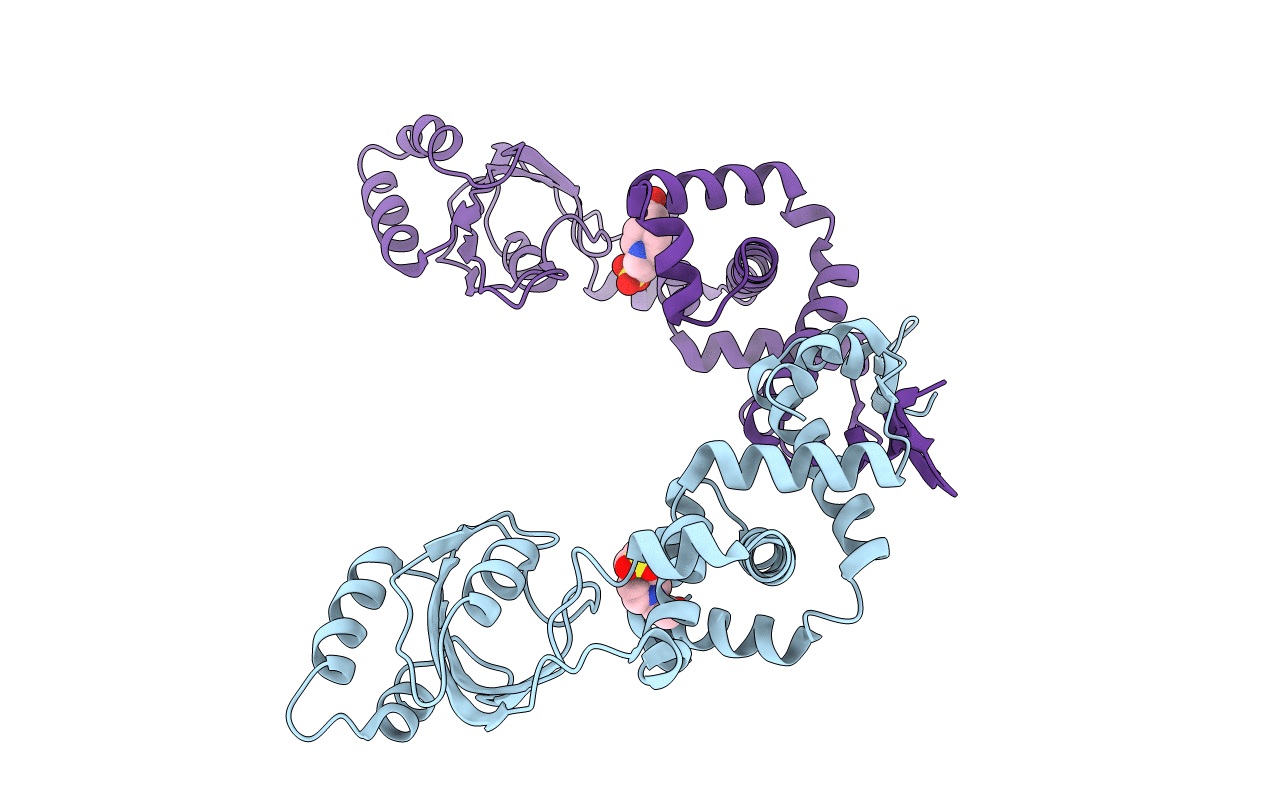
During infection, bacterial pathogens rely on secreted virulence factors to manipulate the host cell. However, in gram-positive bacteria, the molecular mechanisms underlying the folding and activity of these virulence factors after membrane translocation are not clear. Here, we solved the protein structures of two secreted parvulin and two secreted cyclophilin-like peptidyl-prolyl isomerase (PPIase) ATP-independent chaperones found in gram-positive streptococcal species. The extracellular parvulin-type PPIase, PrsA in Streptococcus pneumoniae and Streptococcus mutans maintain dimeric crystal structures reminiscent of folding catalysts that consist of two domains, a PPIase and foldase domain. Structural comparison of the two cyclophilin-like extracellular chaperones from S. pneumoniae and Streptococcus pyogenes with other cyclophilins demonstrates that this group of cyclophilin-like chaperones has novel structural appendages formed by 9- and 24-residue insertions. Furthermore, we demonstrate that deletion of prsA and slrA genes impairs the secretion of the cholesterol-dependent pore-forming toxin, pneumolysin in S. pneumoniae. Using protein pull-down and biophysical assays, we demonstrate a direct interaction between PrsA and SlrA with Ply. Then, we developed chaperone-assisted folding assays that show that the S. pneumoniae PrsA and SlrA extracellular chaperones accelerate pneumolysin folding. In addition, we demonstrate that SlrA and, for the first time, S. pyogenes PpiA exhibit PPIase activity and can bind the immunosuppressive drug, cyclosporine A. Altogether, these findings suggest a mechanistic role for streptococcal PPIase chaperones in the activity and folding of secreted virulence factors such as pneumolysin.
IMPORTANCE
Streptococcal species are a leading cause of lower respiratory infections that annually affect millions of people worldwide. During infection, streptococcal species secrete a medley of virulence factors that allow the bacteria to colonize and translocate to deeper tissues. In many gram-positive bacteria, virulence factors are secreted from the cytosol across the bacterial membrane in an unfolded state. The bacterial membrane-cell wall interface is exposed to the potentially harsh extracellular environment, making it difficult for native virulence factors to fold before being released into the host. ATP-independent PPIase-type chaperones, PrsA and SlrA, are thought to facilitate folding and stabilization of several unfolded proteins to promote the colonization and spread of streptococci. Here, we present crystal structures of the molecular chaperones of PrsA and SlrA homologs from streptococcal species. We provide evidence that the Streptococcus pyogenes SlrA homolog, PpiA, has PPIase activity and binds to cyclosporine A. In addition, we show that Streptococcus pneumoniae PrsA and SlrA directly interact and fold the cholesterol-dependent pore-forming toxin and critical virulence determinant, pneumolysin.