Deposition Date
2020-09-24
Release Date
2020-12-09
Last Version Date
2024-10-09
Entry Detail
PDB ID:
7K80
Keywords:
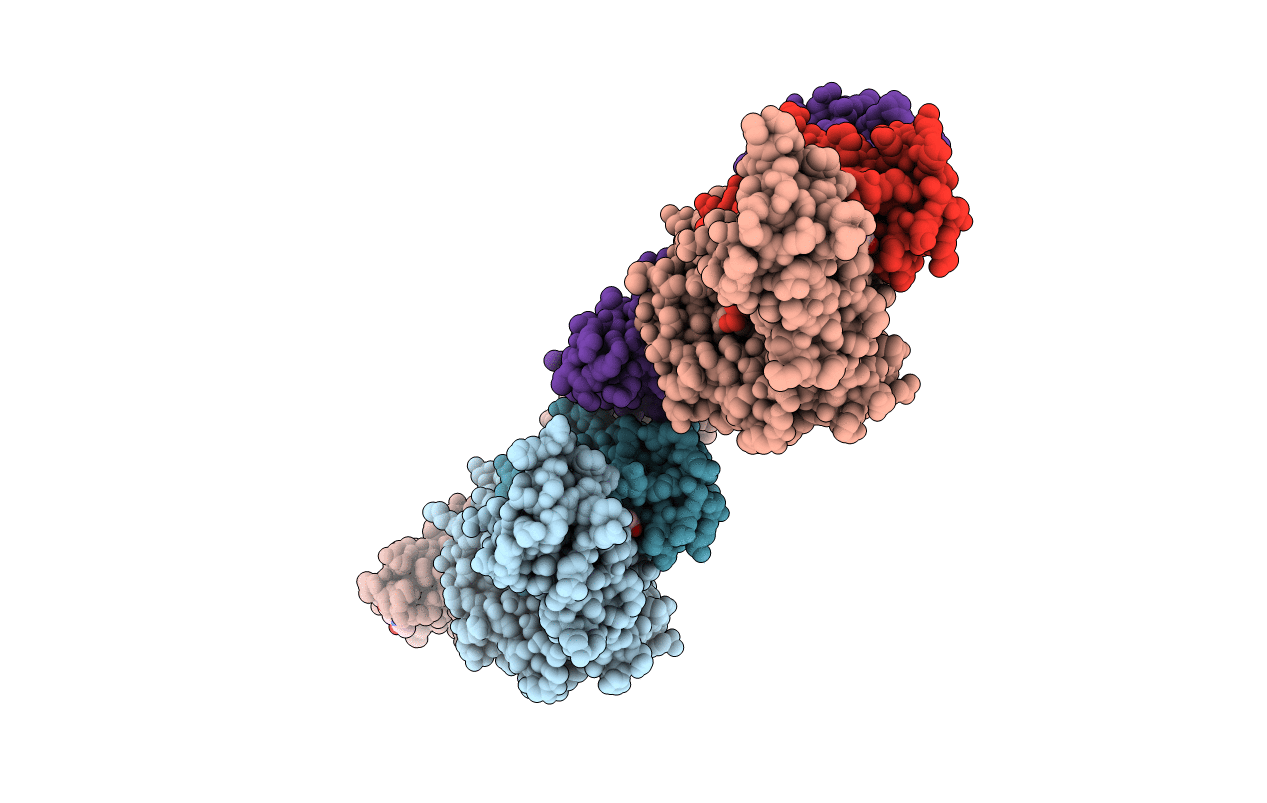
Title:
KIR3DL1*001 in complex with HLA-A*24:02 presenting the RYPLTFGW peptide
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
synthetic construct (Taxon ID: 32630)
synthetic construct (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
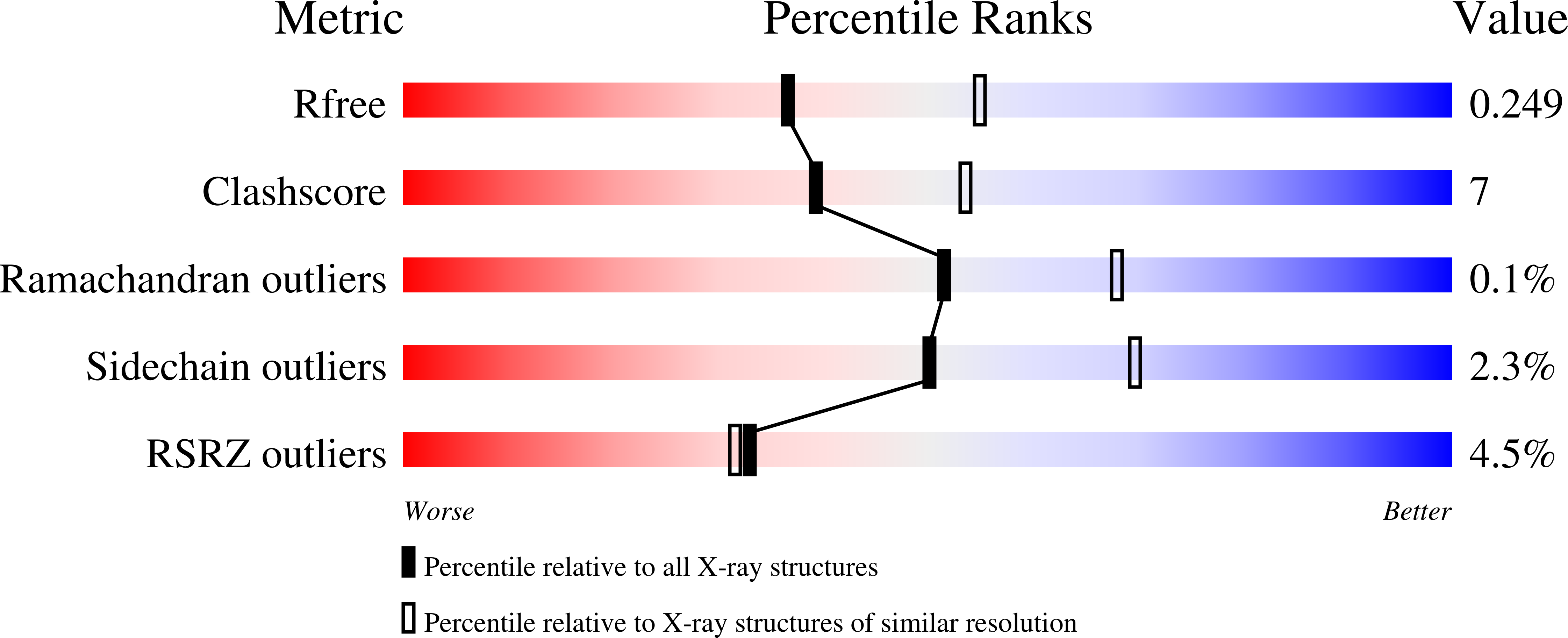
Resolution:
2.40 Å
R-Value Free:
0.24
R-Value Work:
0.19
R-Value Observed:
0.20
Space Group:
P 1