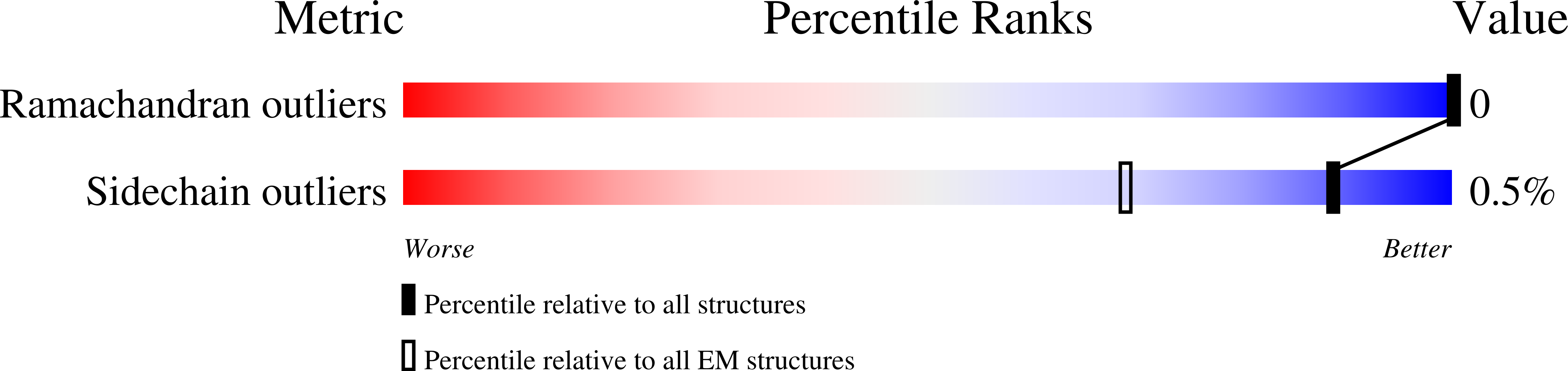Deposition Date
2020-08-24
Release Date
2020-09-16
Last Version Date
2025-05-28
Entry Detail
PDB ID:
7JW1
Keywords:
Title:
Satellite phage P4 procapsid including size determination (Sid) protein
Biological Source:
Source Organism(s):
Escherichia phage P2 (Taxon ID: 10679)
Enterobacteria phage P4 (Taxon ID: 10680)
Enterobacteria phage P4 (Taxon ID: 10680)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
4.19 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE