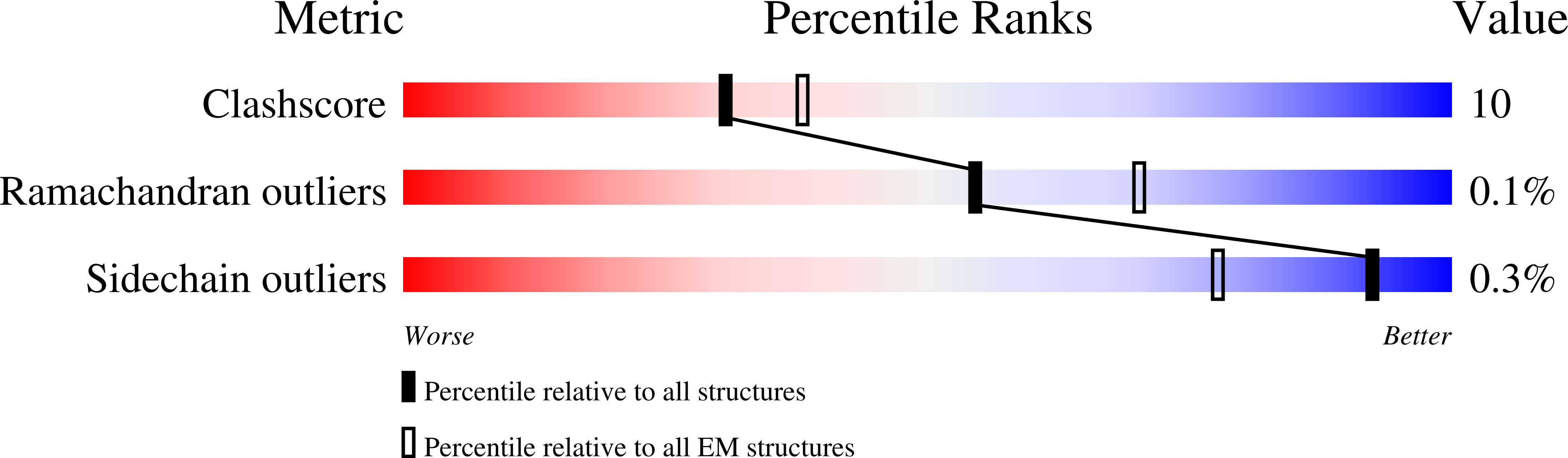
Deposition Date
2020-08-12
Release Date
2021-01-20
Last Version Date
2024-11-20
Entry Detail
PDB ID:
7JRG
Keywords:
Title:
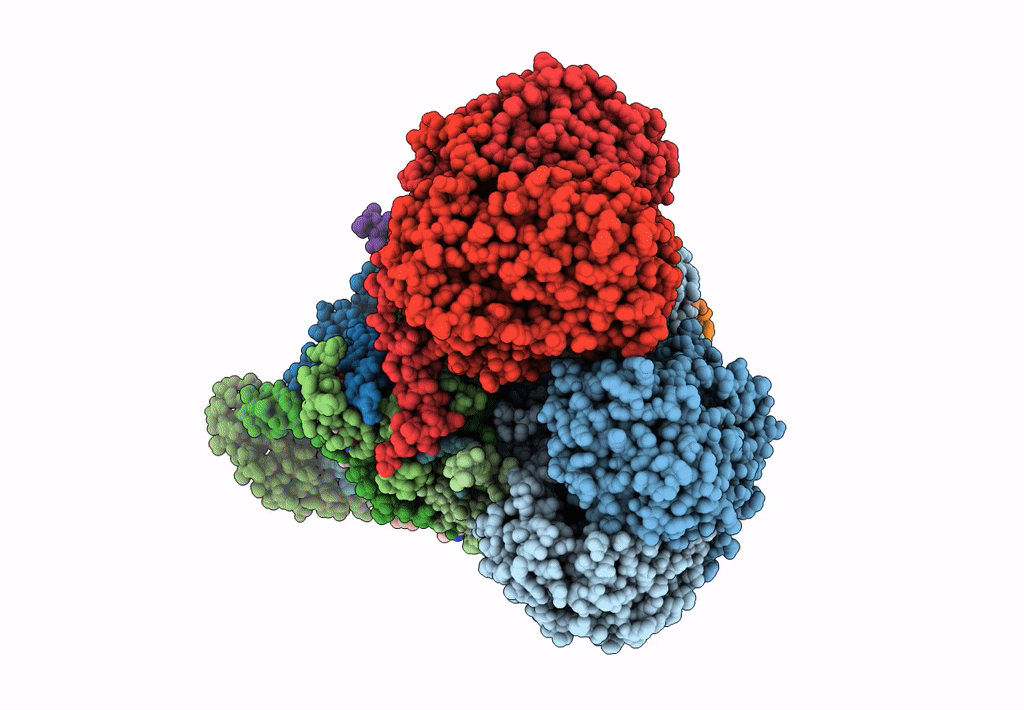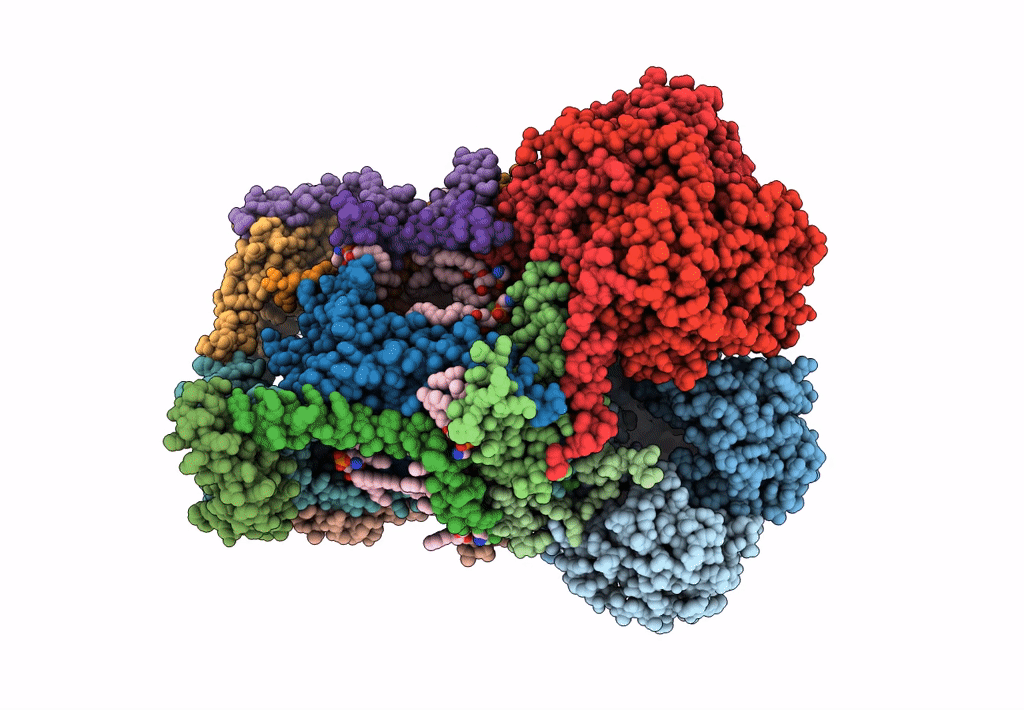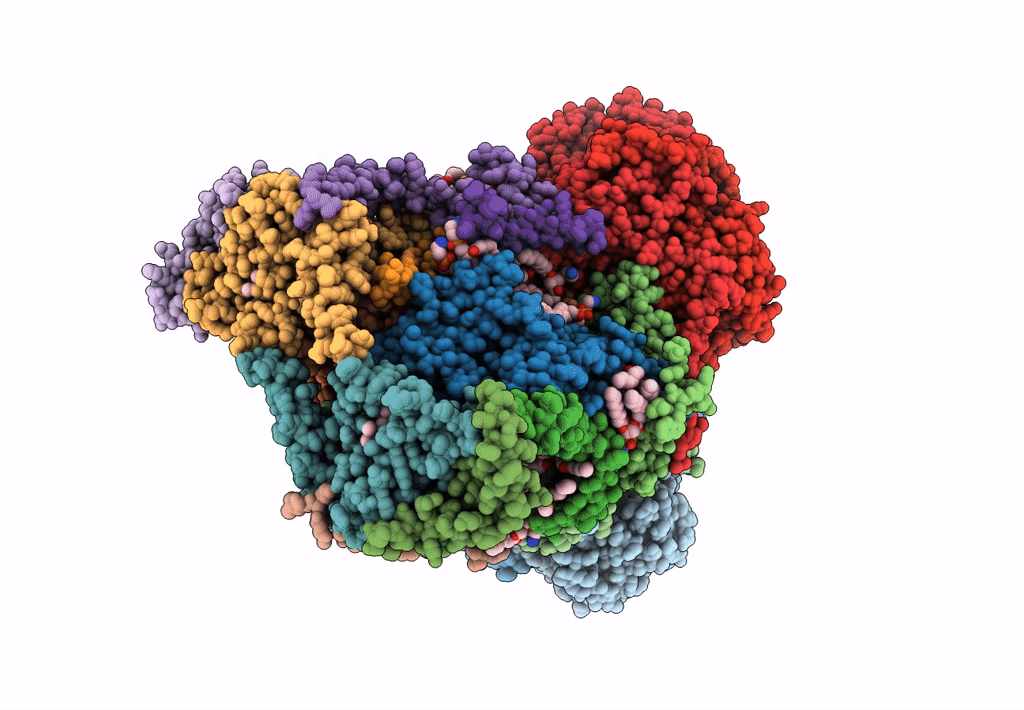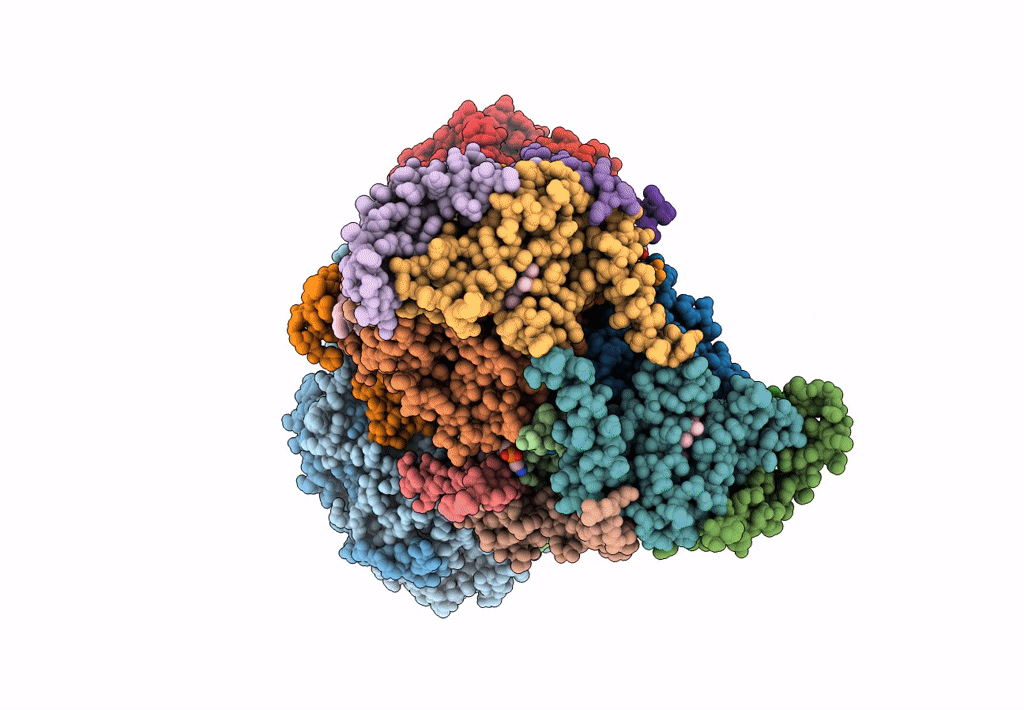
Plant Mitochondrial complex III2 from Vigna radiata
Biological Source:
Source Organism(s):
Vigna radiata var. radiata (Taxon ID: 3916)
Method Details:
Experimental Method:
Resolution:
3.20 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE