Deposition Date
2021-06-17
Release Date
2021-09-22
Last Version Date
2023-11-29
Entry Detail
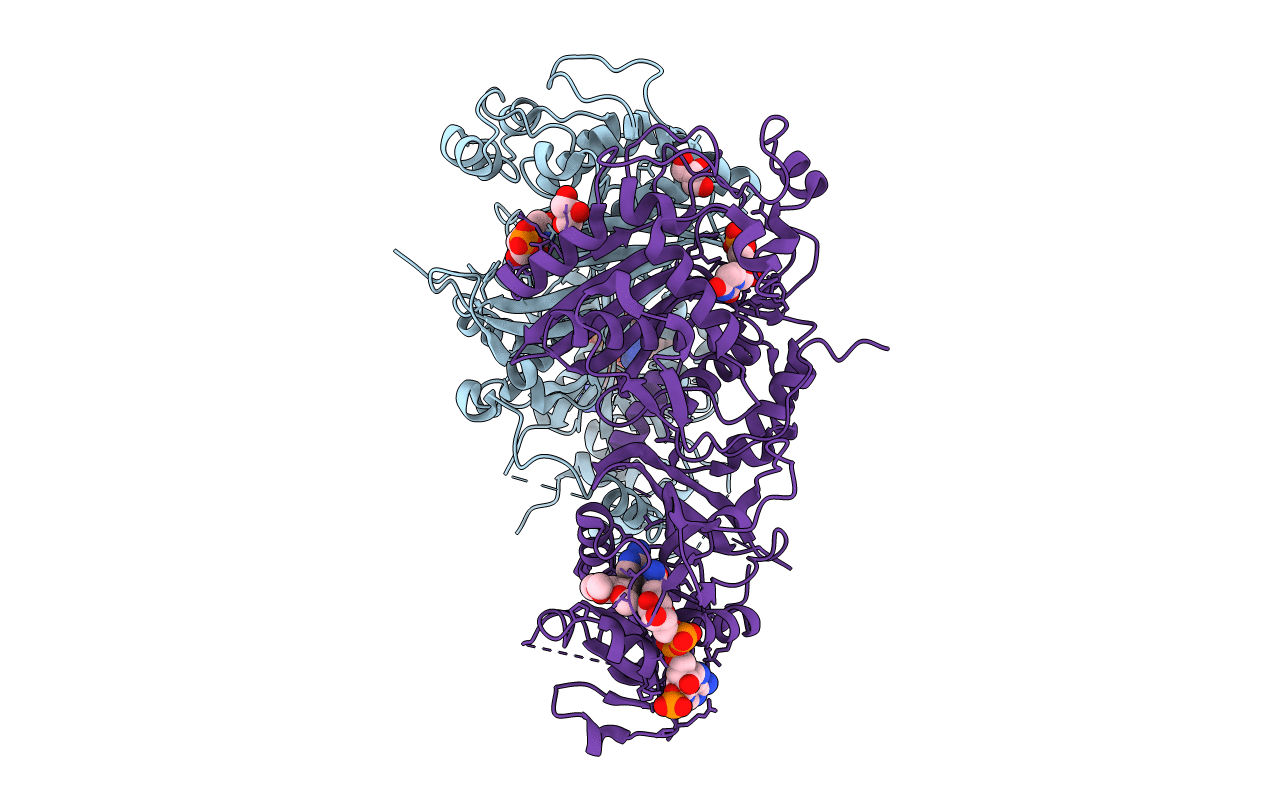
PDB ID:
7F3Z
Keywords:
Title:
Double mutant Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS-K1, C59R+S108N) complexed with Trimethoprim (TOP), NADPH and dUMP
Biological Source:
Source Organism(s):
Plasmodium falciparum (Taxon ID: 5833)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.60 Å
R-Value Free:
0.25
R-Value Work:
0.19
Space Group:
P 21 21 21


