Deposition Date
2021-05-20
Release Date
2021-07-21
Last Version Date
2024-06-05
Entry Detail
PDB ID:
7EV9
Keywords:
Title:
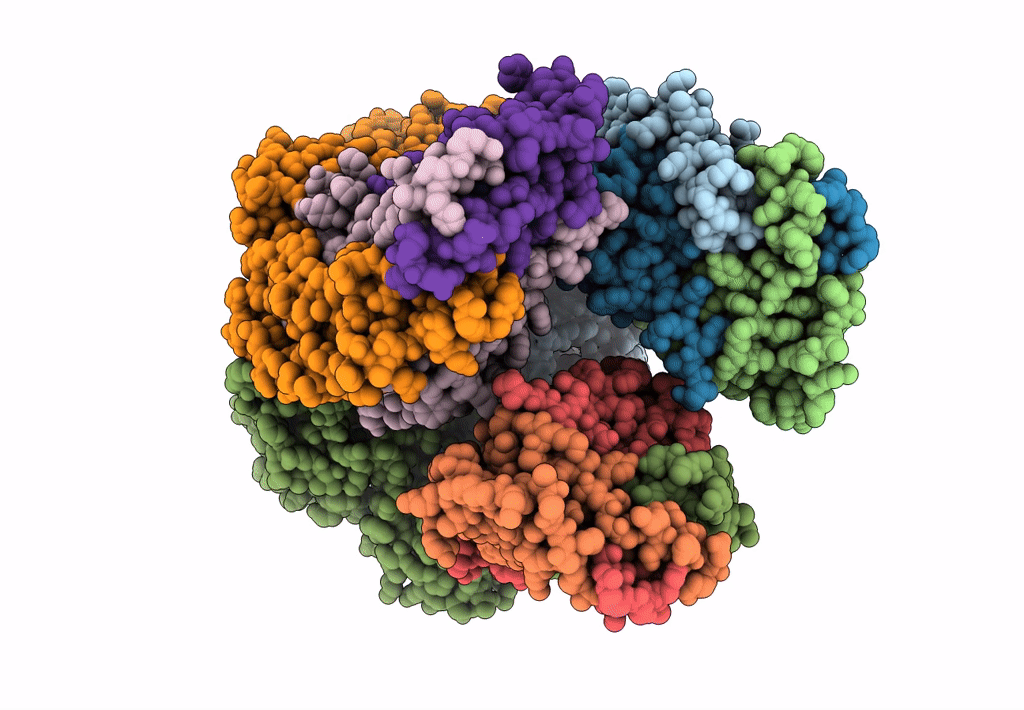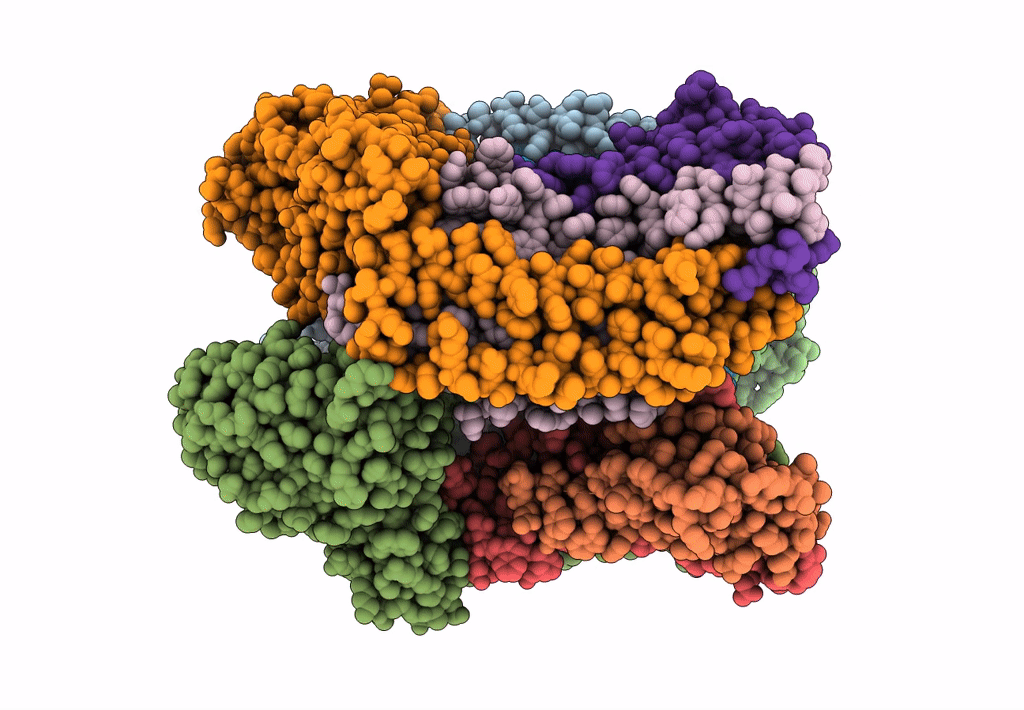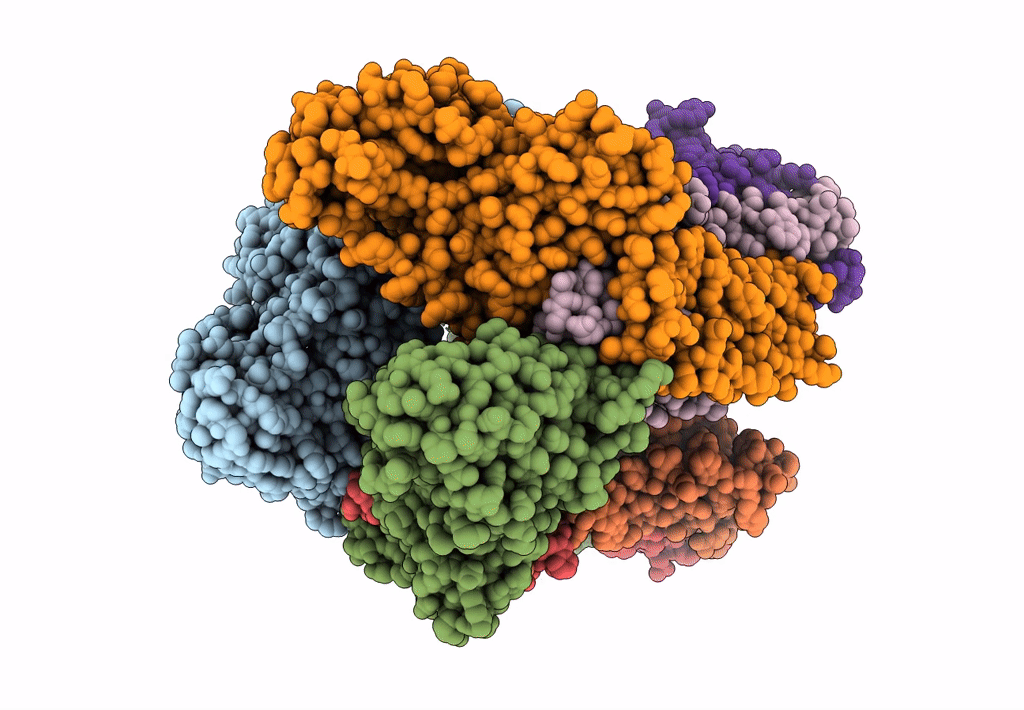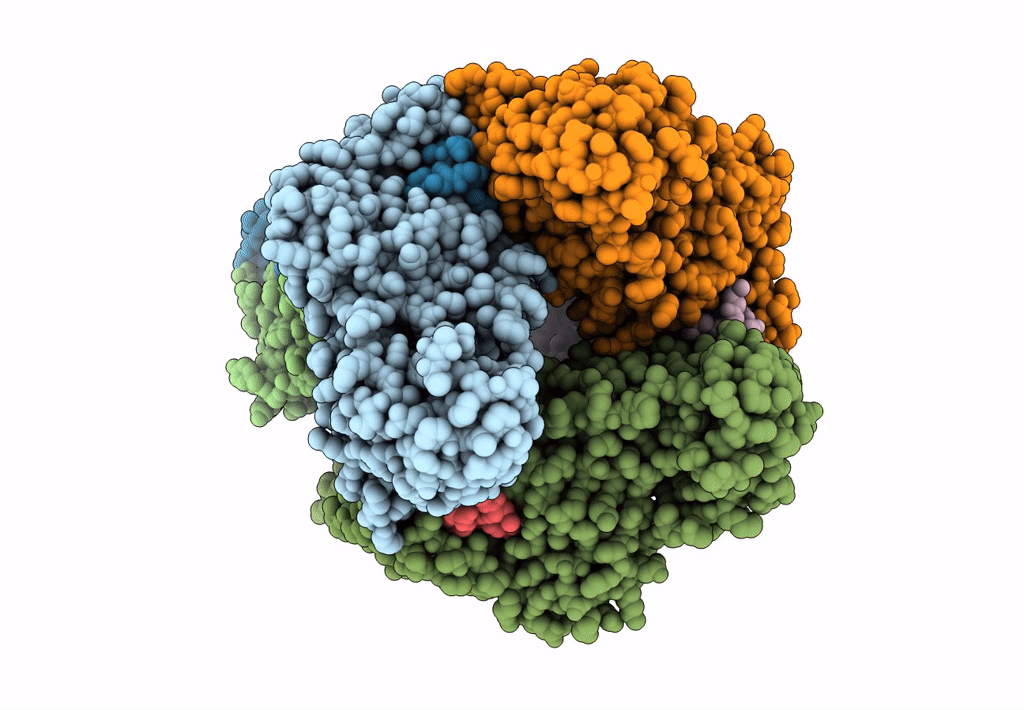
cryoEM structure of particulate methane monooxygenase associated with Cu(I)
Biological Source:
Source Organism(s):
Method Details:
Experimental Method:
Resolution:
2.60 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


