Deposition Date
2021-05-07
Release Date
2022-02-16
Last Version Date
2023-11-29
Entry Detail
PDB ID:
7ERV
Keywords:
Title:
Crystal structure of L-histidine decarboxylase (C57S/C101V/C282V mutant) from Photobacterium phosphoreum
Biological Source:
Source Organism(s):
Photobacterium phosphoreum (Taxon ID: 659)
Expression System(s):
Method Details:
Experimental Method:
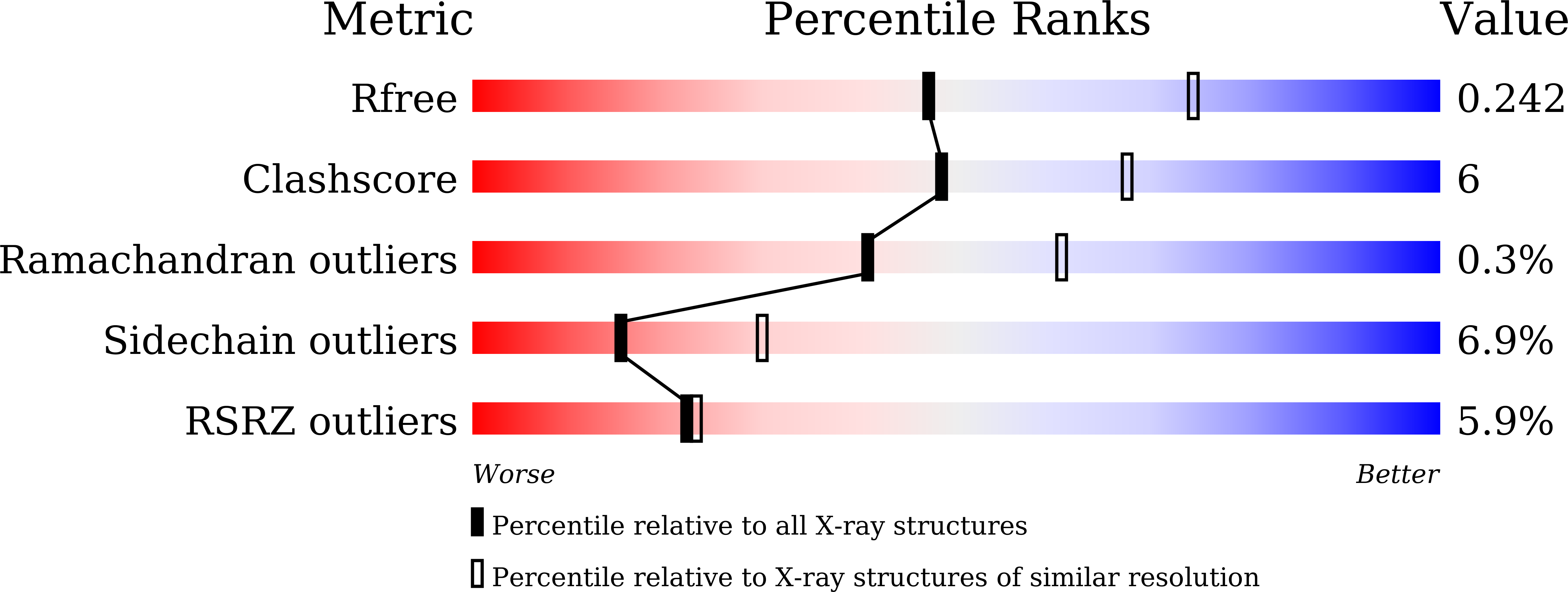
Resolution:
2.50 Å
R-Value Free:
0.23
R-Value Work:
0.18
R-Value Observed:
0.18
Space Group:
P 61 2 2