Deposition Date
2021-04-07
Release Date
2021-11-10
Last Version Date
2023-11-29
Entry Detail
PDB ID:
7EKX
Keywords:
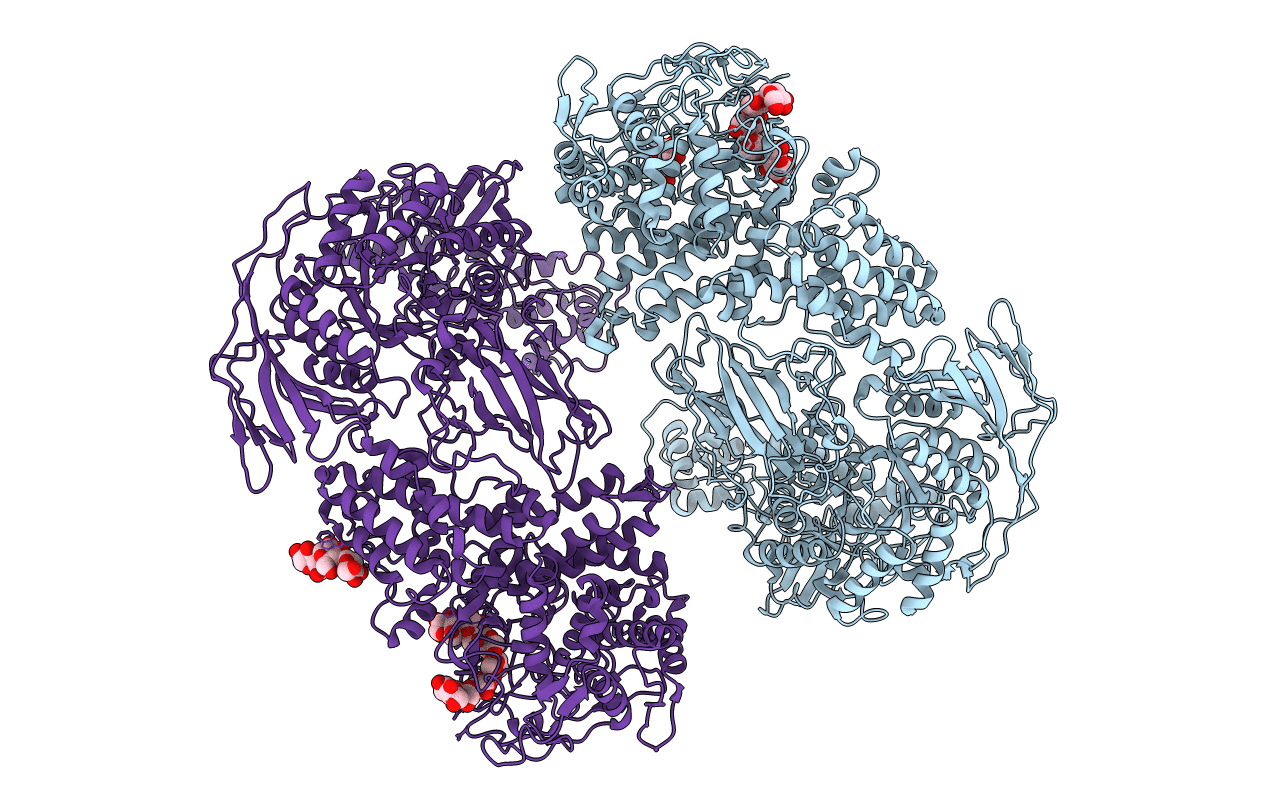
Title:
Crystal Structure of the Candida Glabrata Glycogen Debranching Enzyme (W470A E564Q) in complex with maltononaose
Biological Source:
Source Organism(s):
Candida glabrata CBS 138 (Taxon ID: 284593)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.40 Å
R-Value Free:
0.30
R-Value Work:
0.27
R-Value Observed:
0.27
Space Group:
C 2 2 21


