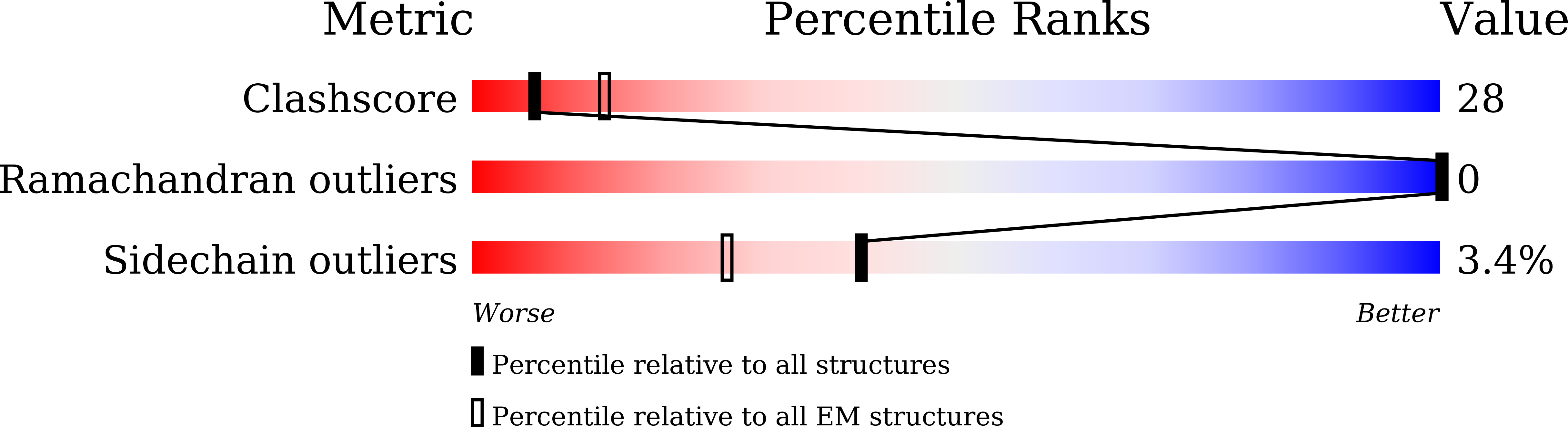
Deposition Date
2021-03-05
Release Date
2021-11-17
Last Version Date
2024-10-16
Entry Detail
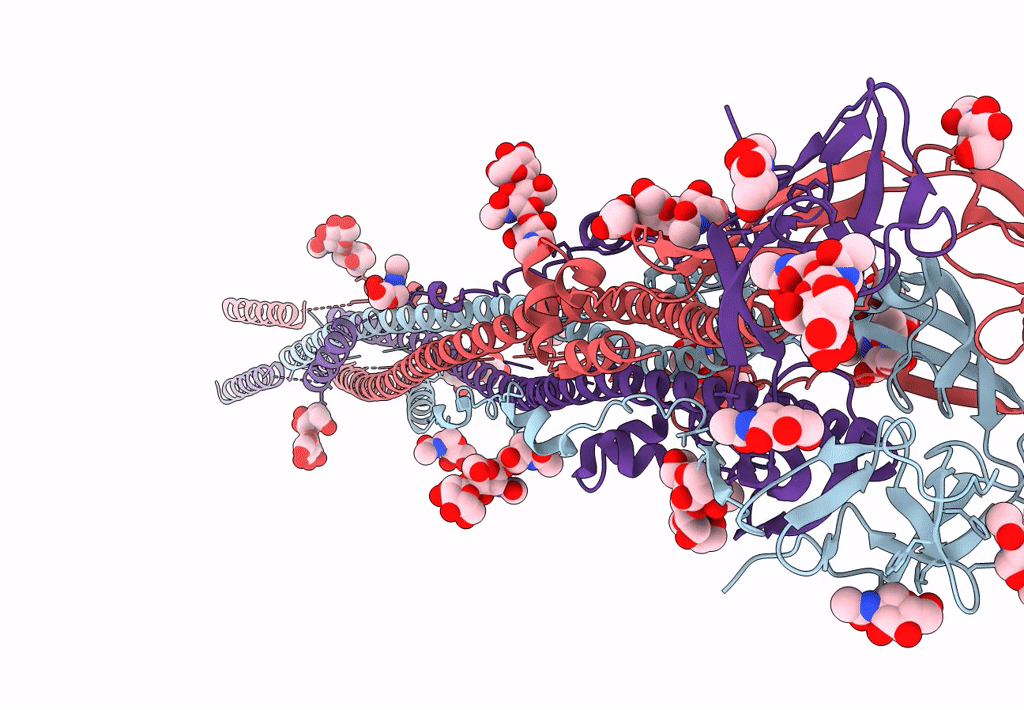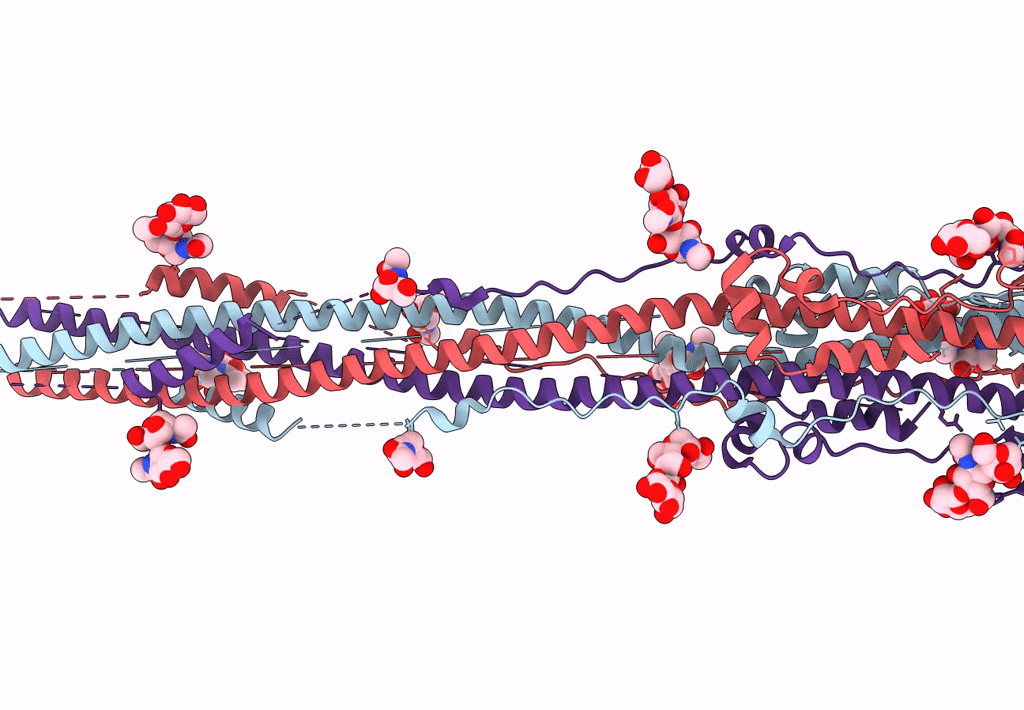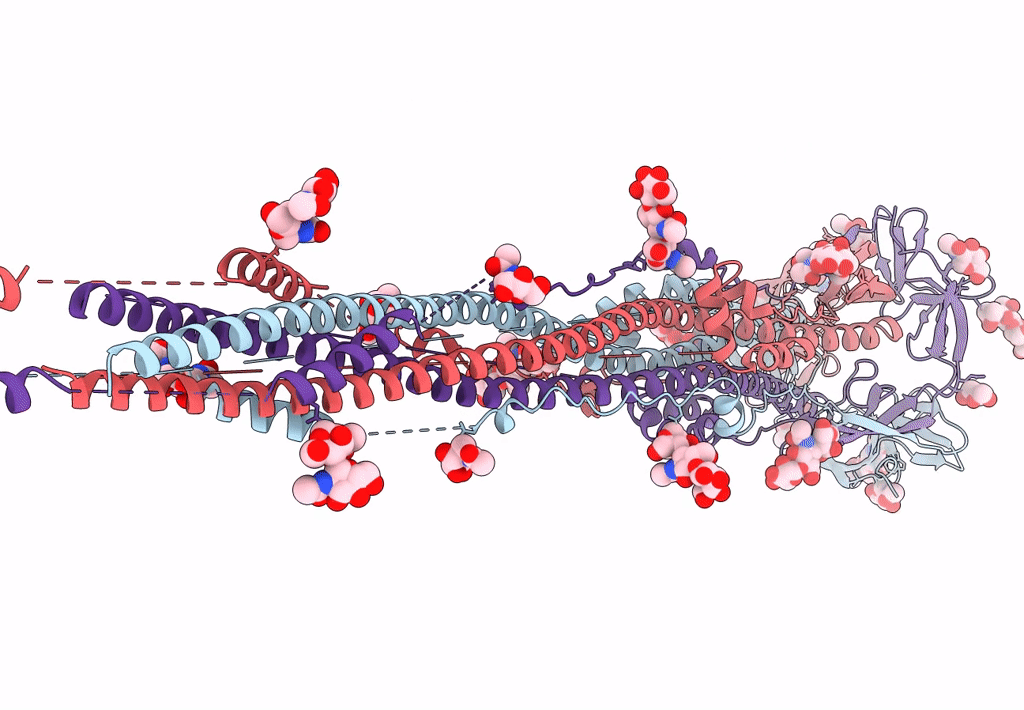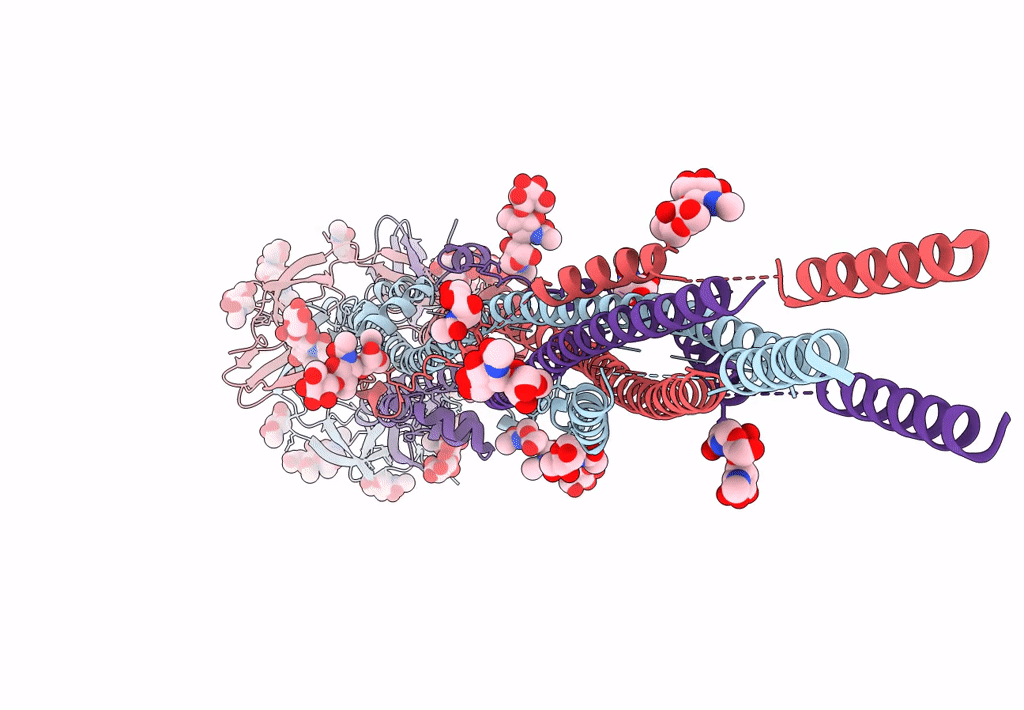
PDB ID:
7E9T
Keywords:
Title:
Nanometer resolution in situ structure of SARS-CoV-2 post-fusion spike
Biological Source:
Source Organism(s):
Method Details:
Experimental Method:
Resolution:
10.90 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SUBTOMOGRAM AVERAGING