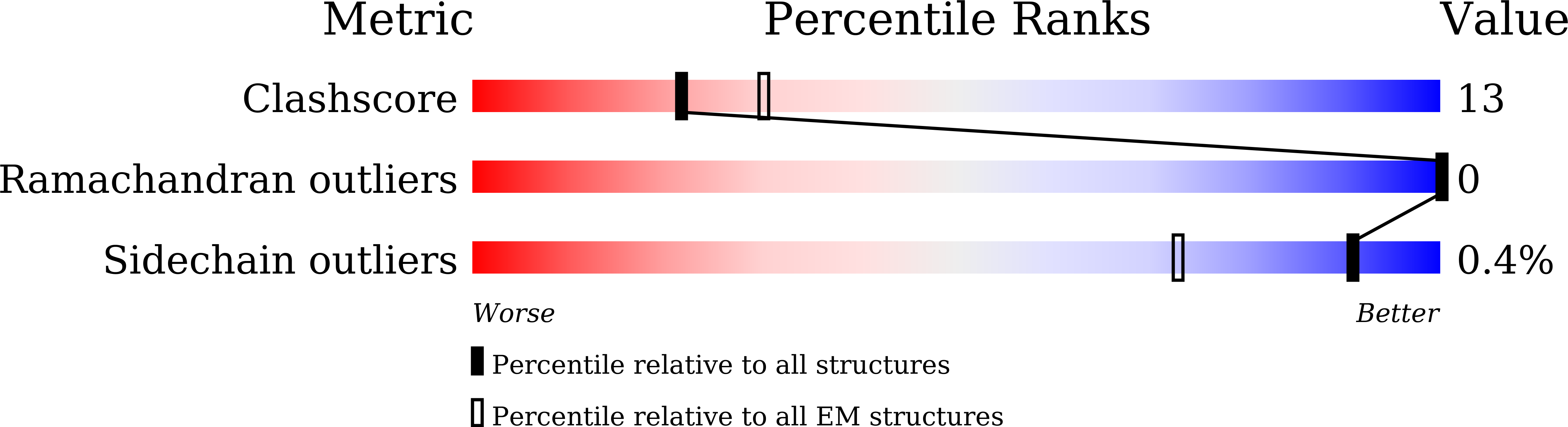
Deposition Date
2021-01-17
Release Date
2021-06-02
Last Version Date
2025-07-02
Entry Detail
PDB ID:
7DWQ
Keywords:
Title:
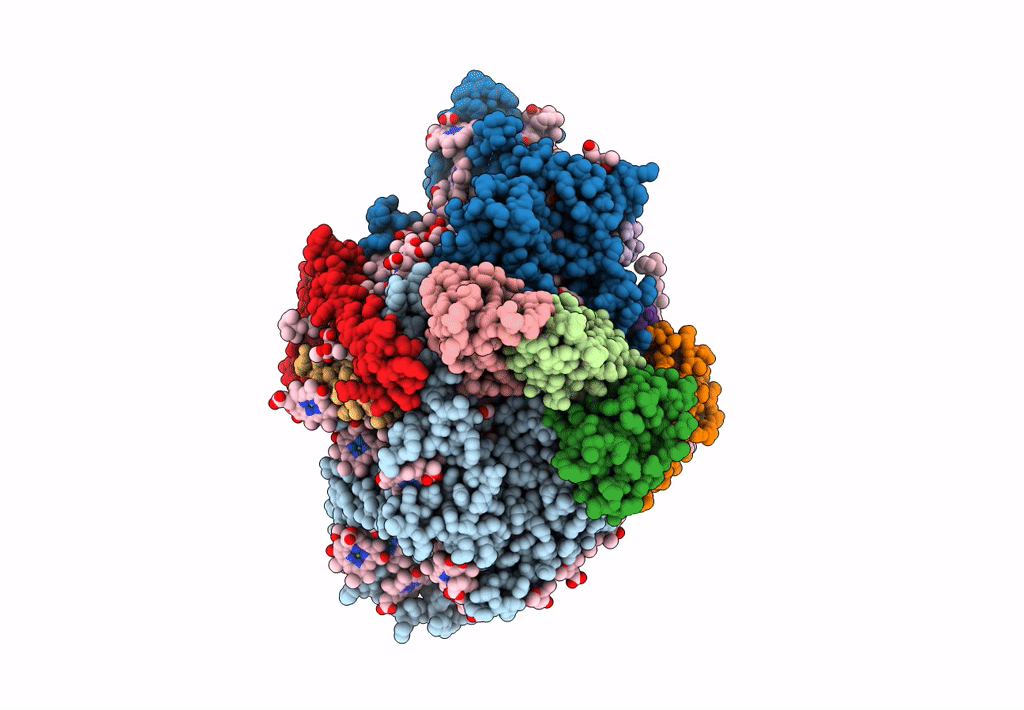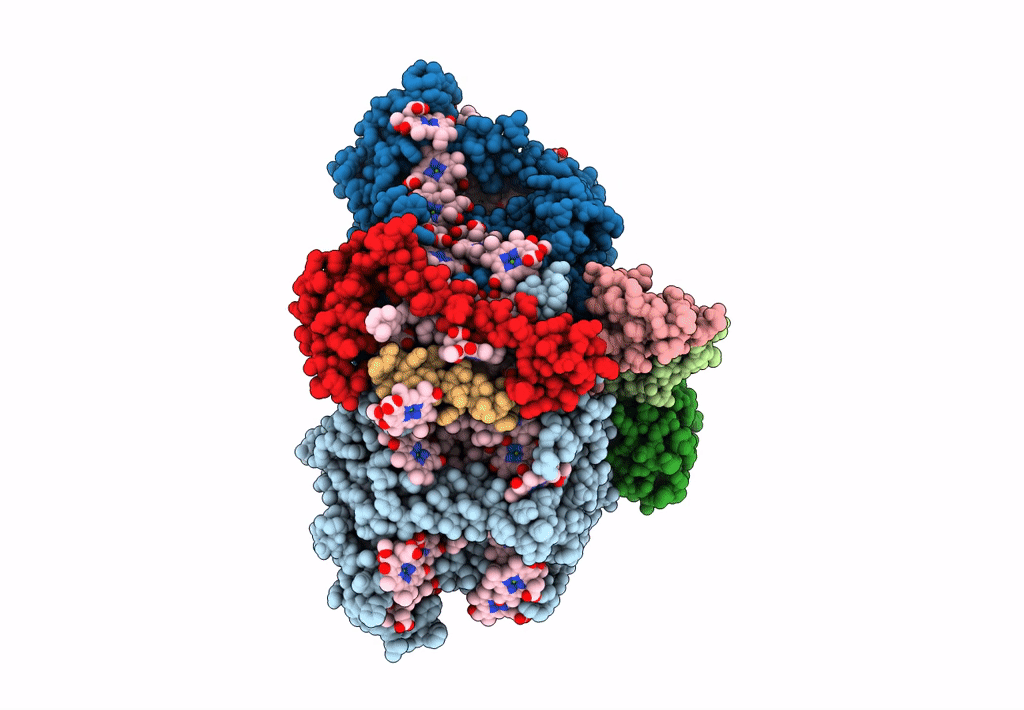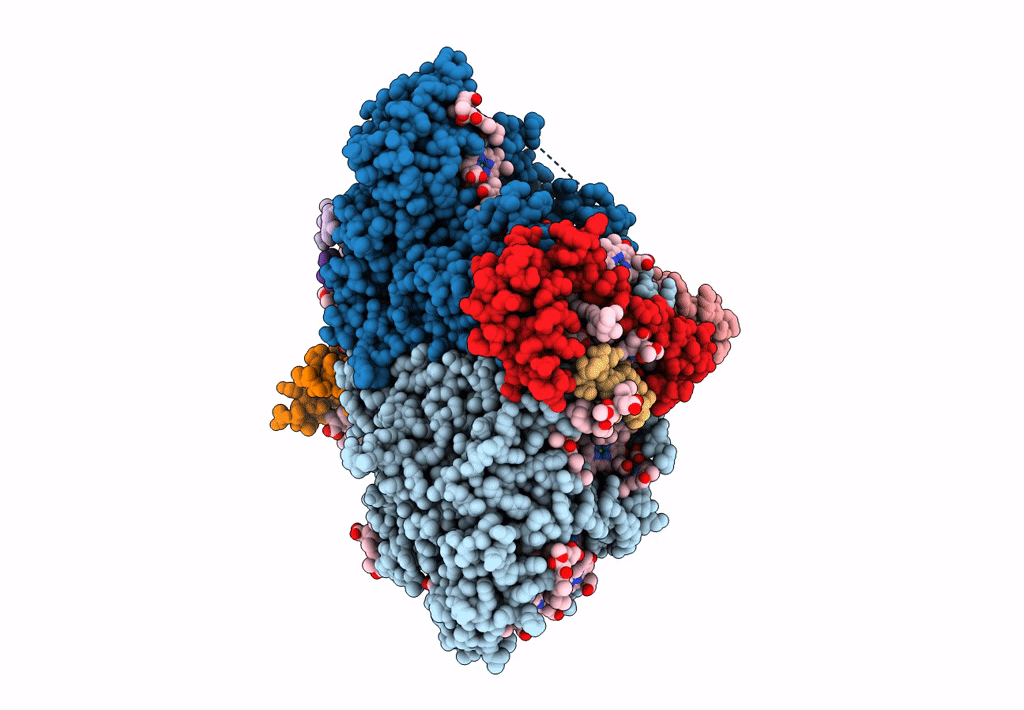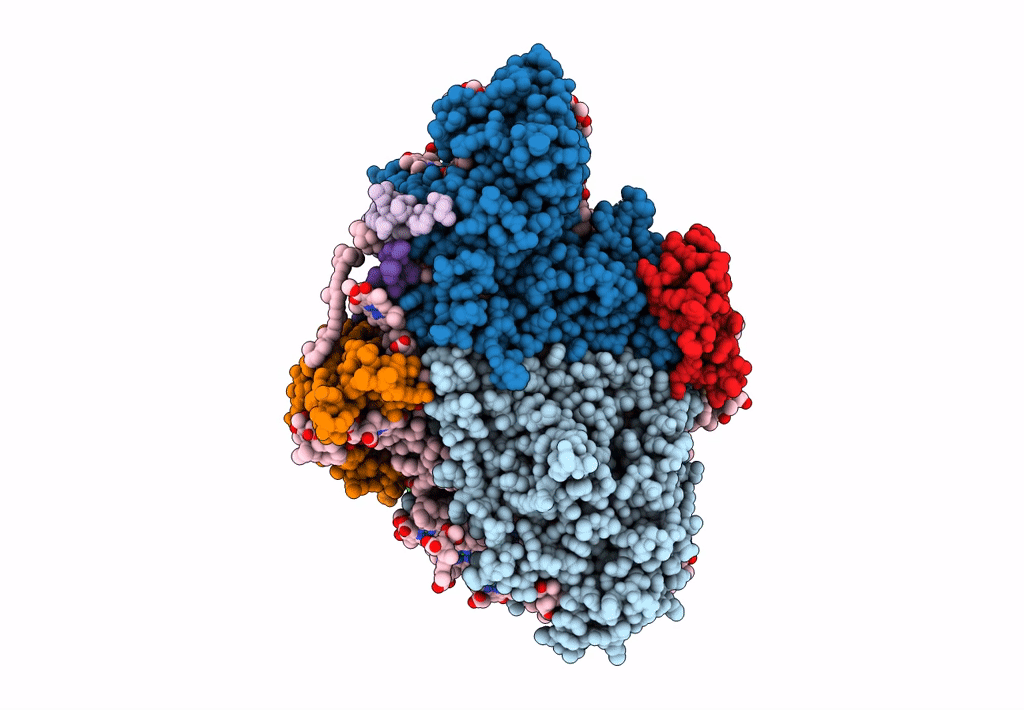
Photosystem I from a chlorophyll d-containing cyanobacterium Acaryochloris marina
Biological Source:
Source Organism(s):
Acaryochloris marina MBIC11017 (Taxon ID: 329726)
Method Details:
Experimental Method:
Resolution:
3.30 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE