Deposition Date
2020-12-29
Release Date
2021-09-08
Last Version Date
2023-11-29
Entry Detail
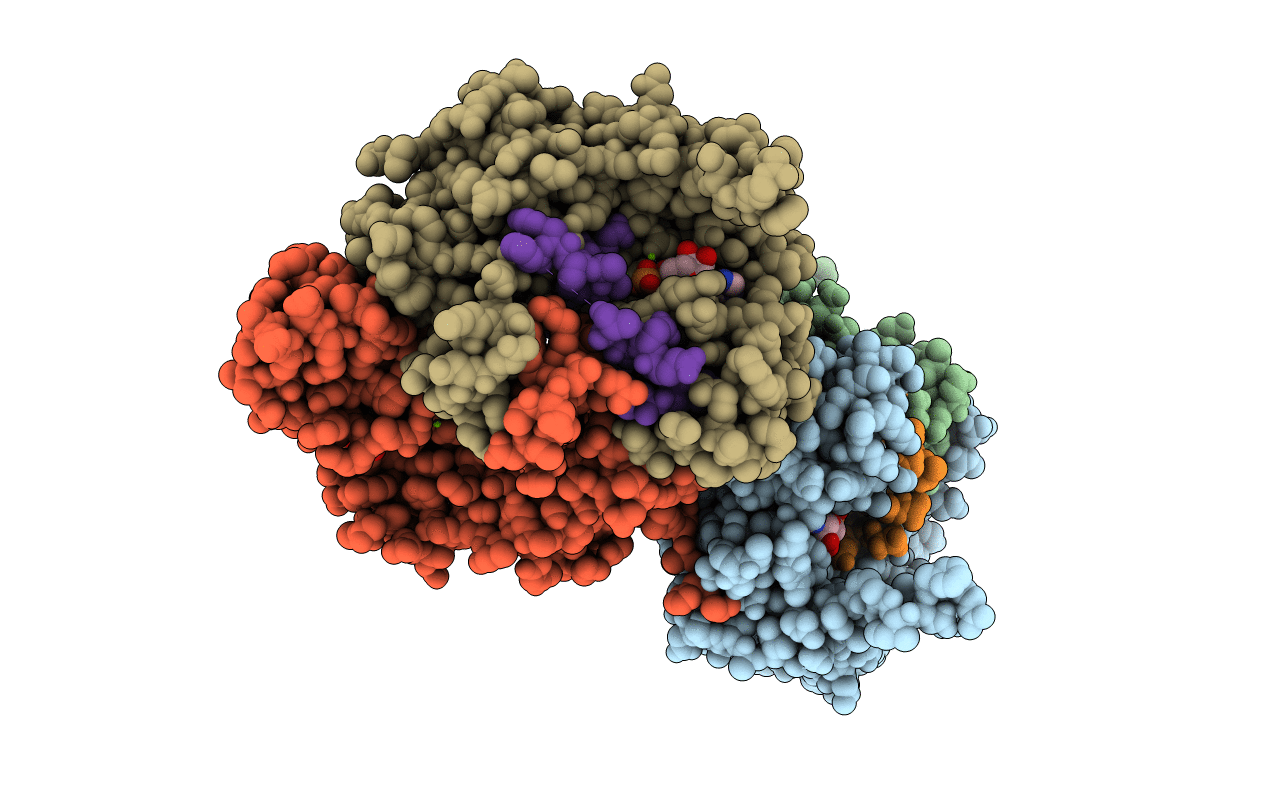
PDB ID:
7DRP
Keywords:
Title:
Structure of ATP-grasp ligase PsnB complexed with phosphomimetic variant of minimal precursor, Mg, and ADP
Biological Source:
Source Organism(s):
Plesiocystis pacifica SIR-1 (Taxon ID: 391625)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.98 Å
R-Value Free:
0.20
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
P 1 21 1


