Deposition Date
2020-08-20
Release Date
2021-02-03
Last Version Date
2024-11-06
Entry Detail
PDB ID:
7CTS
Keywords:
Title:
Open form of PET-degrading cutinase Cut190 with thermostability-improving mutations of S226P/R228S/Q138A/D250C-E296C/Q123H/N202H and S176A inactivation
Biological Source:
Source Organism(s):
Saccharomonospora viridis (Taxon ID: 1852)
Expression System(s):
Method Details:
Experimental Method:
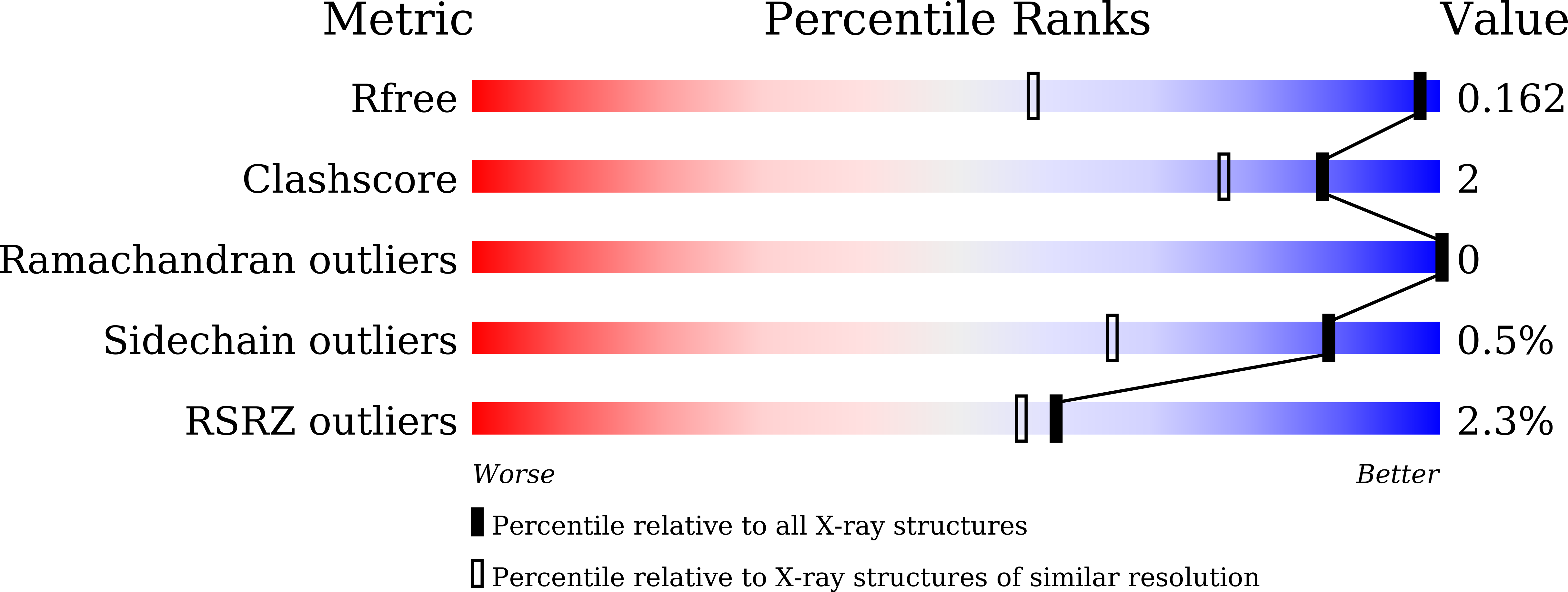
Resolution:
1.10 Å
R-Value Free:
0.16
R-Value Work:
0.14
R-Value Observed:
0.14
Space Group:
P 21 21 21