Abstact
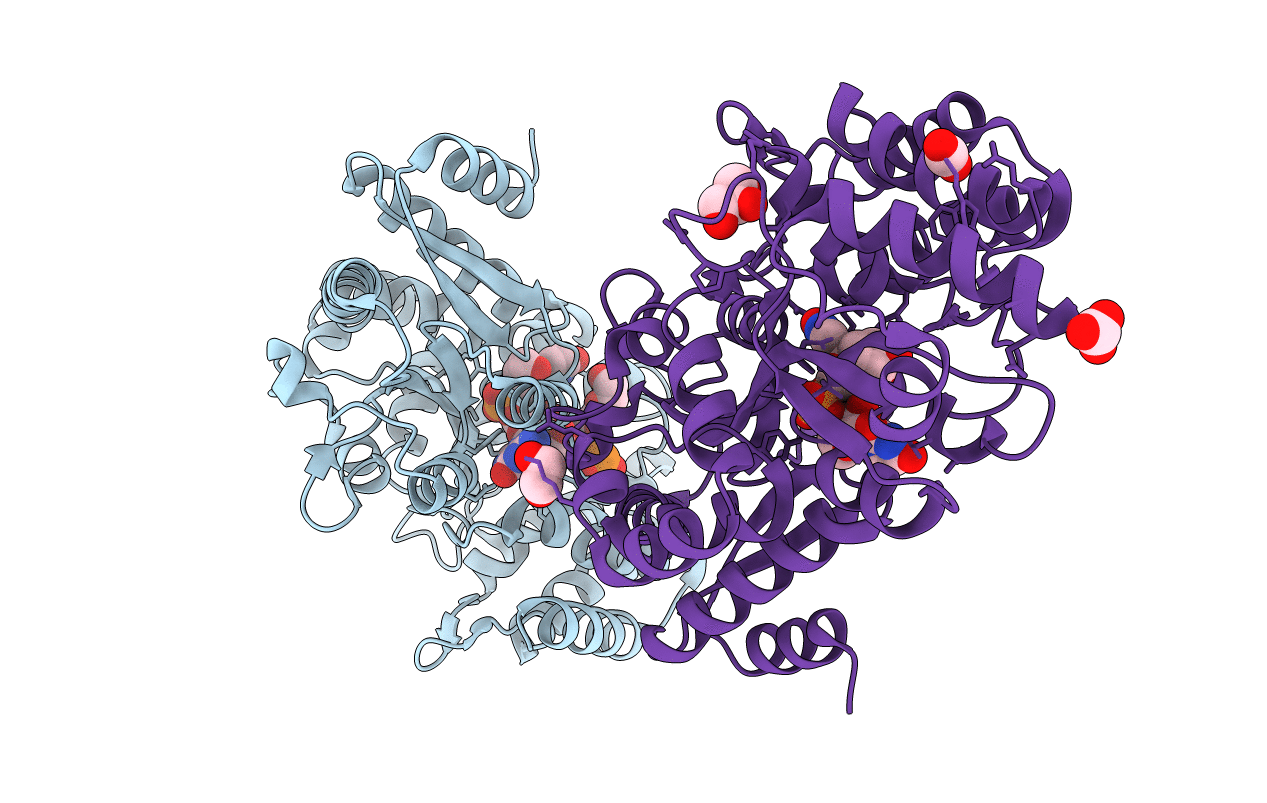
Substrate (or solute)-binding proteins (SBPs) selectively bind the target ligands and deliver them to the ATP-binding cassette (ABC) transport system for their translocation. Irrespective of the different types of ligands, SBPs are structurally conserved. A wealth of structural details of SBPs bound to different types of ligands and the physiological basis of their import are available; however, the uptake mechanism of nucleotides is still deficient. In this study, we elucidated the structural details of an SBP endogenously bound to a novel ligand, a derivative of uridylyl-3'-5'-phospho-guanosine (U3G); thus, we named it a U3G-binding protein (U3GBP). To the best of our knowledge, this is the first report of U3G (and a dinucleotide) binding to the SBP of ABC transport system, and thus, U3GBP is classified as a first member of subcluster D-I SBPs. Thermodynamic data also suggest that U3GBP can bind phospholipid precursor sn-glycerophosphocholine (GPC) at a site other than the active site. Moreover, a combination of mutagenic and structural information reveals that the protein U3GBP follows the well-known 'Venus Fly-trap' mechanism for dinucleotide binding. DATABASES: Structural data are available in RCSB Protein Data Bank under the accession number(s) 7C0F, 7C0K, 7C0L, 7C0O, 7C0R, 7C0S, 7C0T, 7C0U, 7C0V, 7C0W, 7C0X, 7C0Y, 7C0Z, 7C14, 7C15, 7C16, 7C19, and 7C1B.