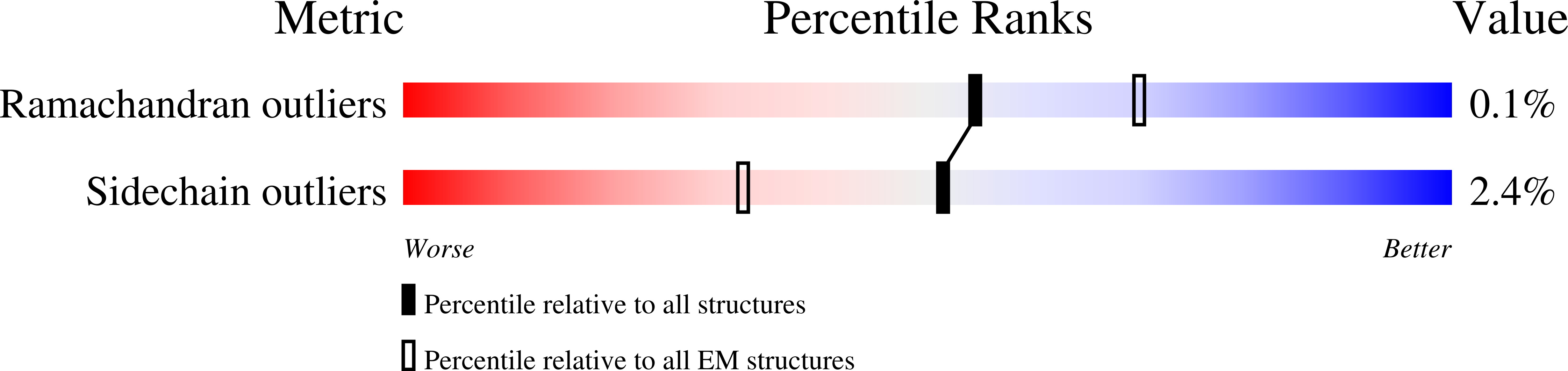
Deposition Date
2021-01-15
Release Date
2021-09-29
Last Version Date
2025-07-02
Entry Detail
PDB ID:
7BKE
Keywords:
Title:
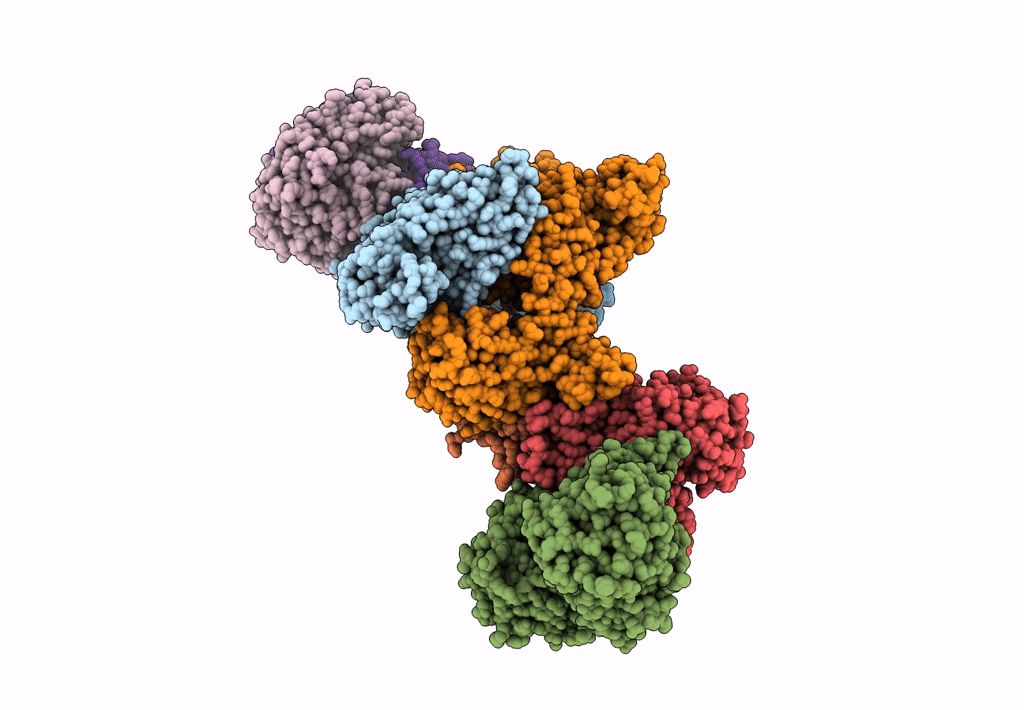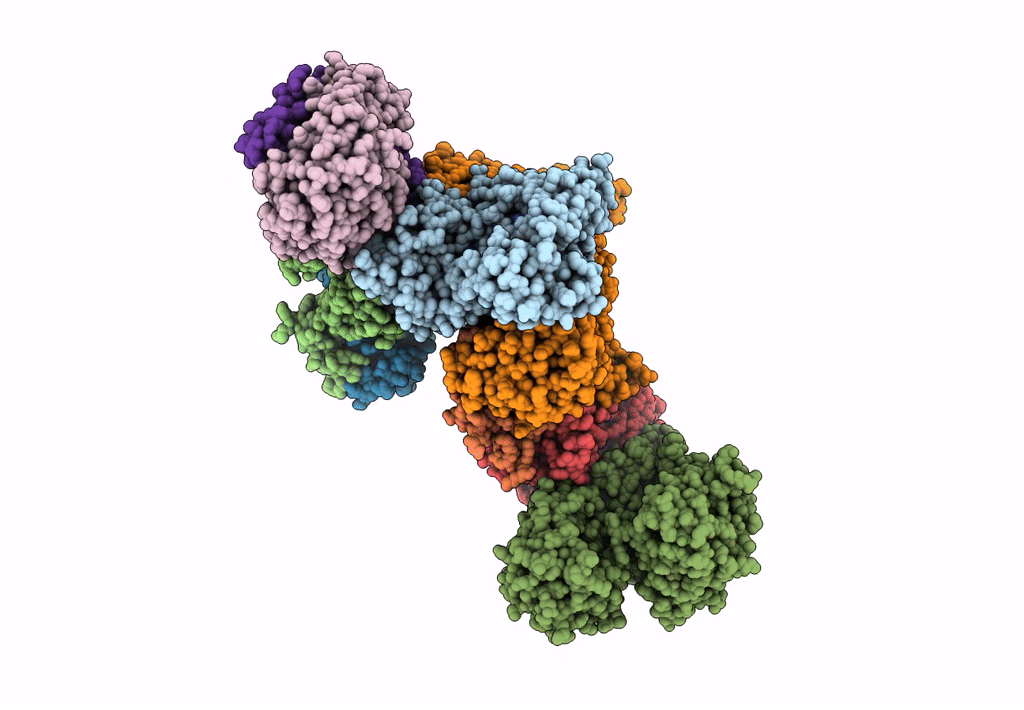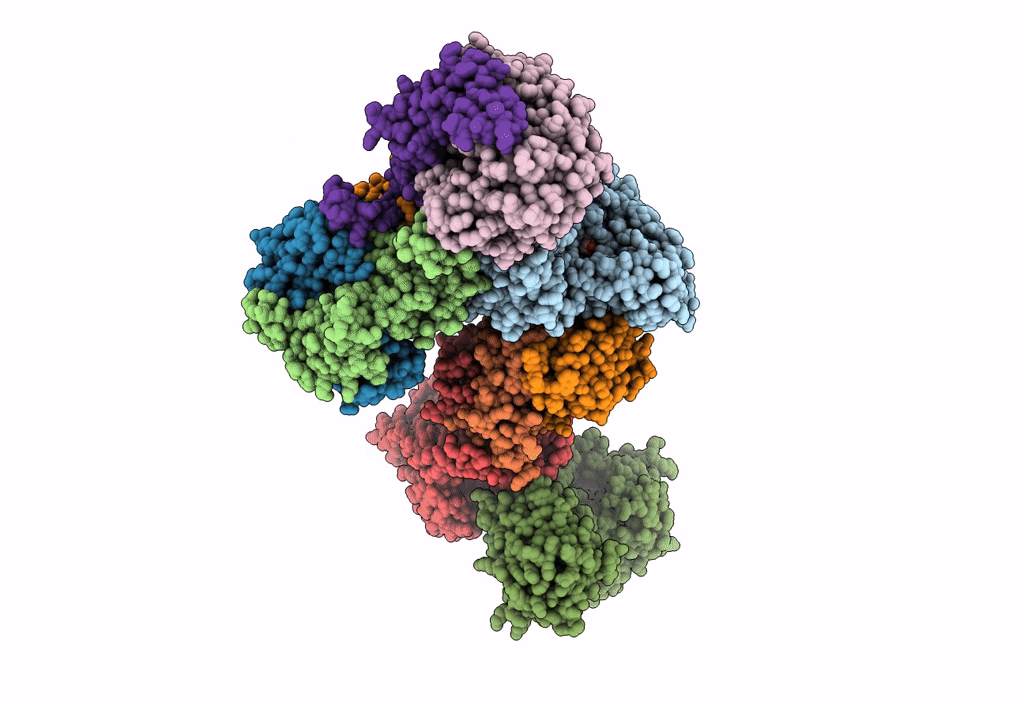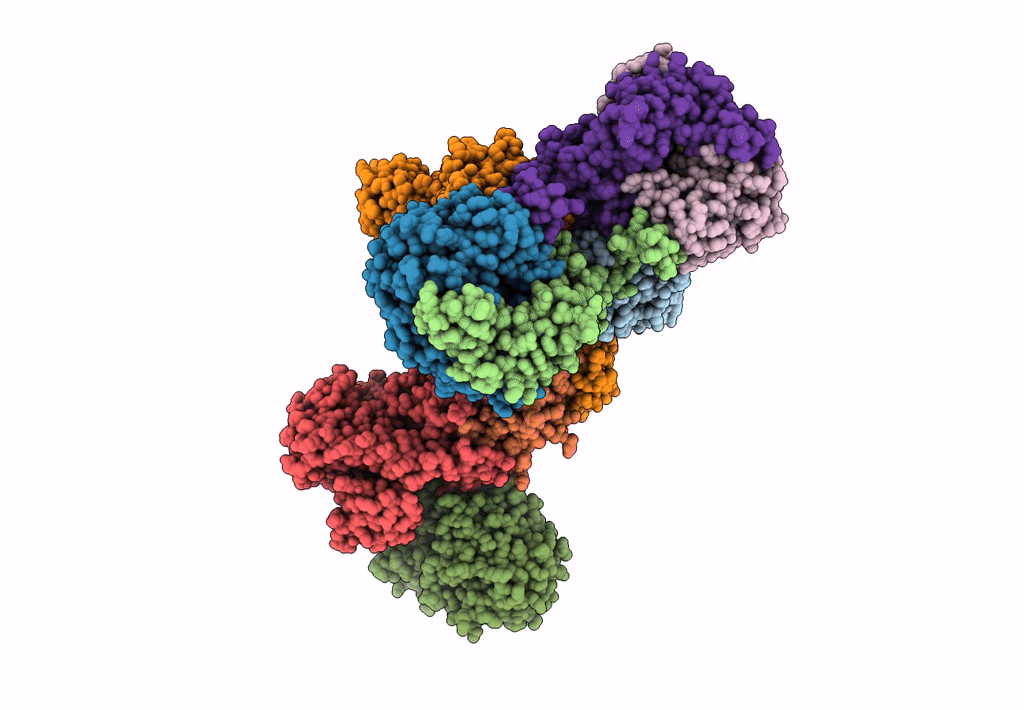
Formate dehydrogenase - heterodisulfide reductase - formylmethanofuran dehydrogenase complex from Methanospirillum hungatei (heterodisulfide reductase core and mobile arm in conformational state 2, composite structure)
Biological Source:
Source Organism(s):
Methanospirillum hungatei JF-1 (Taxon ID: 323259)
Method Details:
Experimental Method:
Resolution:
2.80 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE