Deposition Date
2020-11-23
Release Date
2021-12-01
Last Version Date
2024-11-06
Entry Detail
PDB ID:
7B12
Keywords:
Title:
HUMAN IMMUNOPROTEASOME 20S PARTICLE IN COMPLEX WITH [2-(3-ethylphenyl)-1-[(2S)-3-phenyl-2-[(pyrazin-2-yl)formamido]propanamido]ethyl]boronic acid
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Method Details:
Experimental Method:
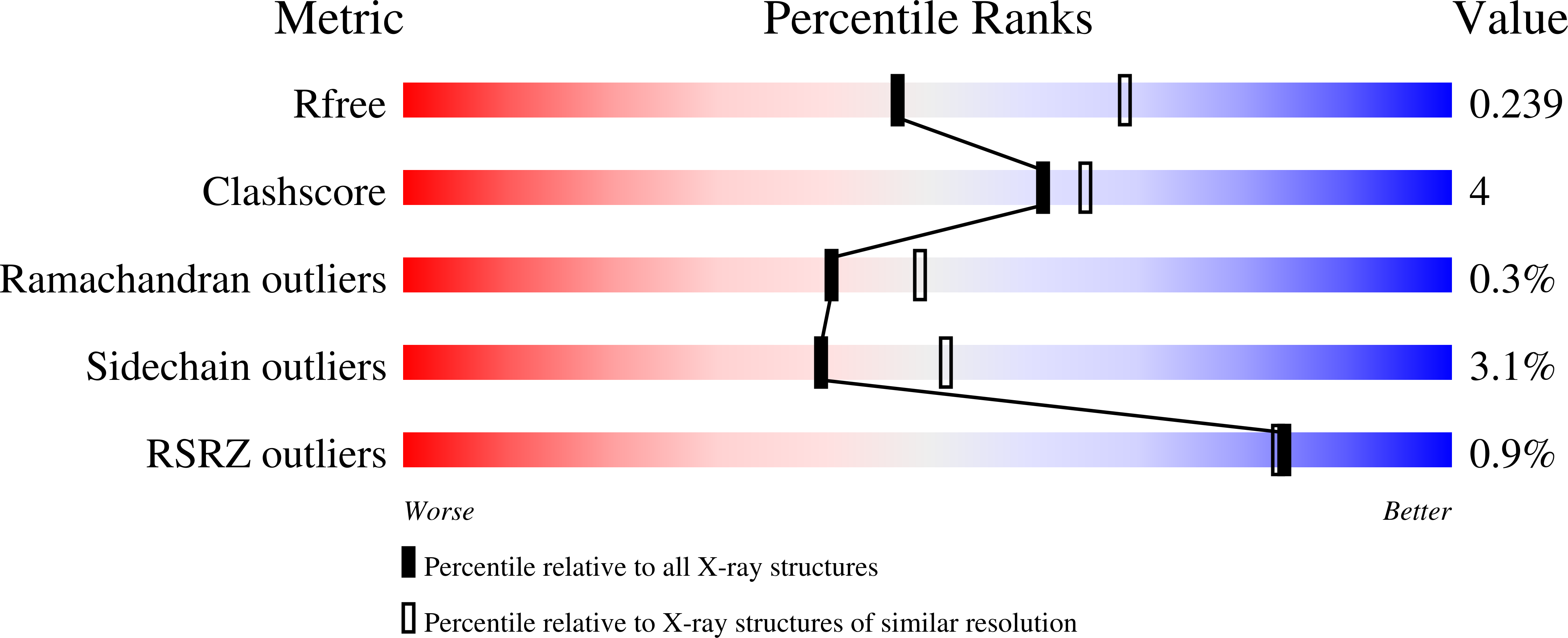
Resolution:
2.43 Å
R-Value Free:
0.22
R-Value Work:
0.18
R-Value Observed:
0.18
Space Group:
P 1 21 1