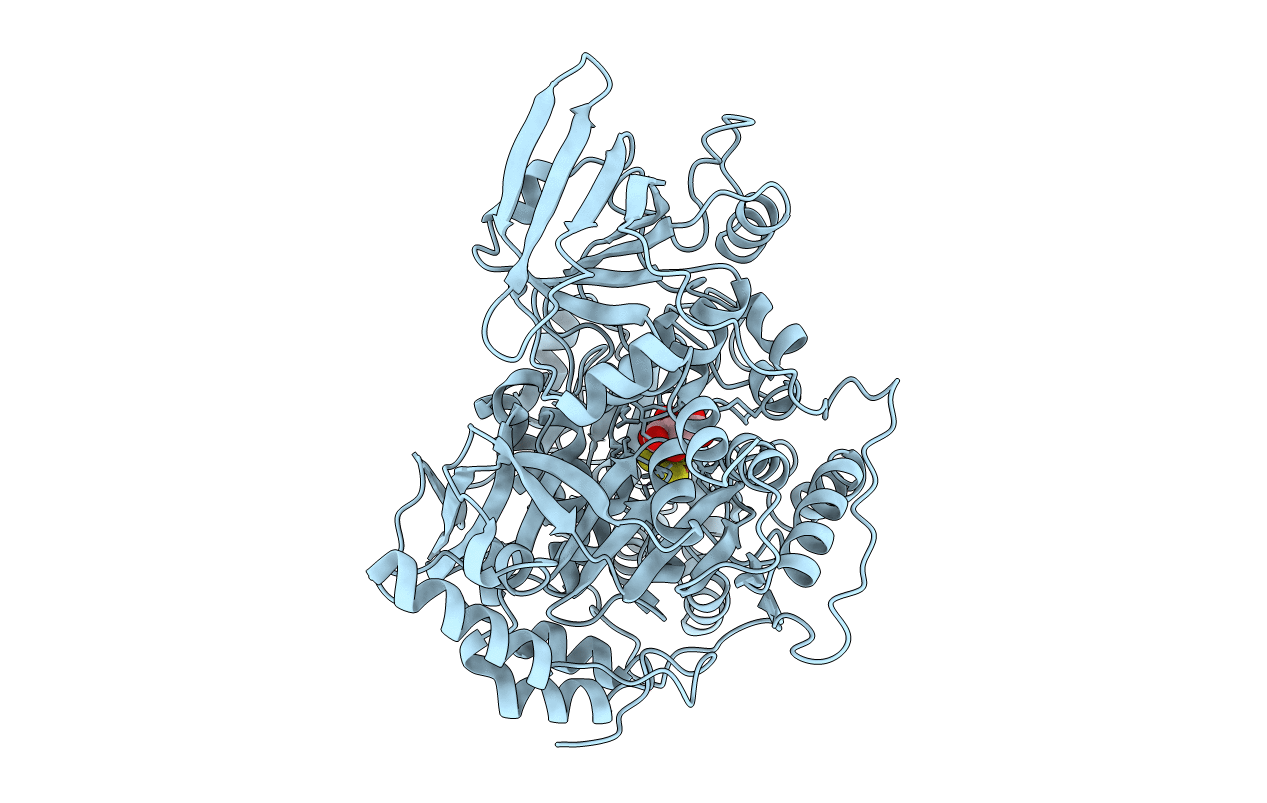Abstact
The crystal structures of mitochondrial aconitase with isocitrate and nitroisocitrate bound have been solved and refined to R factors of 0.179 and 0.161, respectively, for all observed data in the range 8.0-2.1 A. Porcine heart enzyme was used for determining the structure with isocitrate bound. The presence of isocitrate in the crystals was corroborated by Mössbauer spectroscopy. Bovine heart enzyme was used for determining the structure with the reaction intermediate analogue nitroisocitrate bound. The inhibitor binds to the enzyme in a manner virtually identical to that of isocitrate. Both compounds bind to the unique Fe atom of the [4Fe-4S] cluster via a hydroxyl oxygen and one carboxyl oxygen. A H2O molecule is also bound, making Fe six-coordinate. The unique Fe is pulled away approximately 0.2 A from the corner of the cubane compared to the position it would occupy in a symmetrically ligated [4Fe-4S] cluster. At least 23 residues from all four domains of aconitase contribute to the active site. These residues participate in substrate recognition (Arg447, Arg452, Arg580, Arg644, Gln72, Ser166, Ser643), cluster ligation and interaction (Cys358, Cys421, Cys424, Asn258, Asn446), and hydrogen bonds supporting active site side chains (Ala74, Asp568, Ser571, Thr567). Residues implicated in catalysis are Ser642 and three histidine-carboxylate pairs (Asp100-His101, Asp165-His147, Glu262-His167). The base necessary for proton abstraction from C beta of isocitrate appears to be Ser642; the O gamma atom is proximal to the calculated hydrogen position, while the environment of O gamma suggests stabilization of an alkoxide (an oxyanion hole formed by the amide and side chain of Arg644). The histidine-carboxylate pairs appear to be required for proton transfer reactions involving two oxygens bound to Fe, one derived from solvent (bound H2O) and one derived from substrate hydroxyl. Each oxygen is in contact with a histidine, and both are in contact with the side chain of Asp165, which bridges the two sites on the six-coordinate Fe.