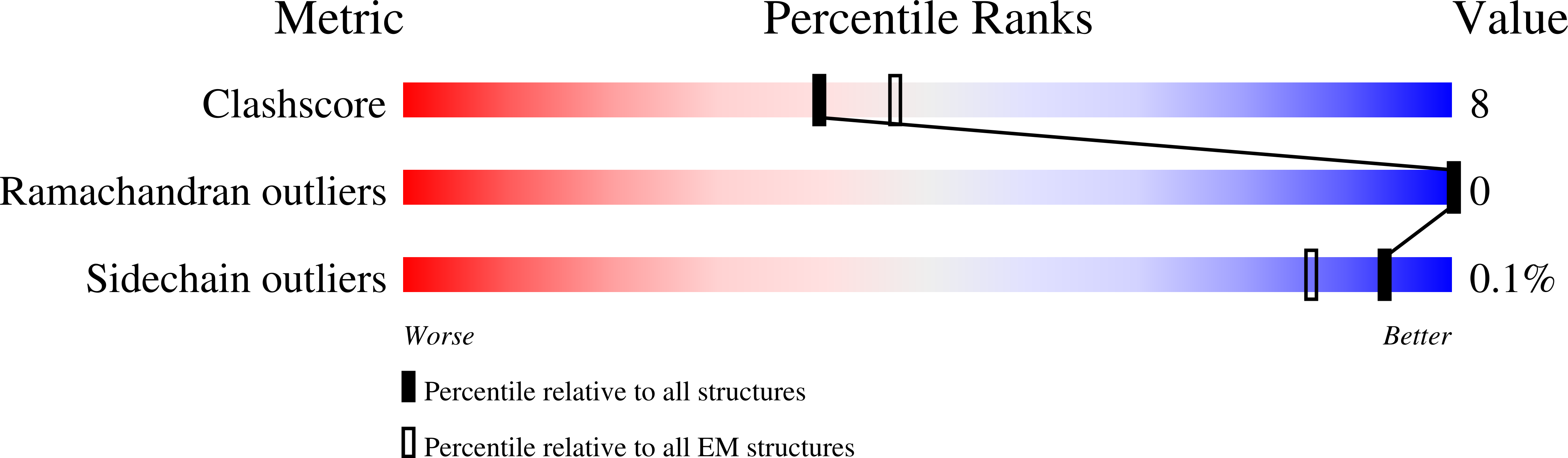Deposition Date
2020-08-12
Release Date
2021-06-09
Last Version Date
2025-05-14
Entry Detail
PDB ID:
7A1D
Keywords:
Title:
Cryo-EM map of the large glutamate dehydrogenase composed of 180 kDa subunits from Mycobacterium smegmatis (open conformation)
Biological Source:
Source Organism(s):
Mycolicibacterium smegmatis MC2 155 (Taxon ID: 246196)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
4.19 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE