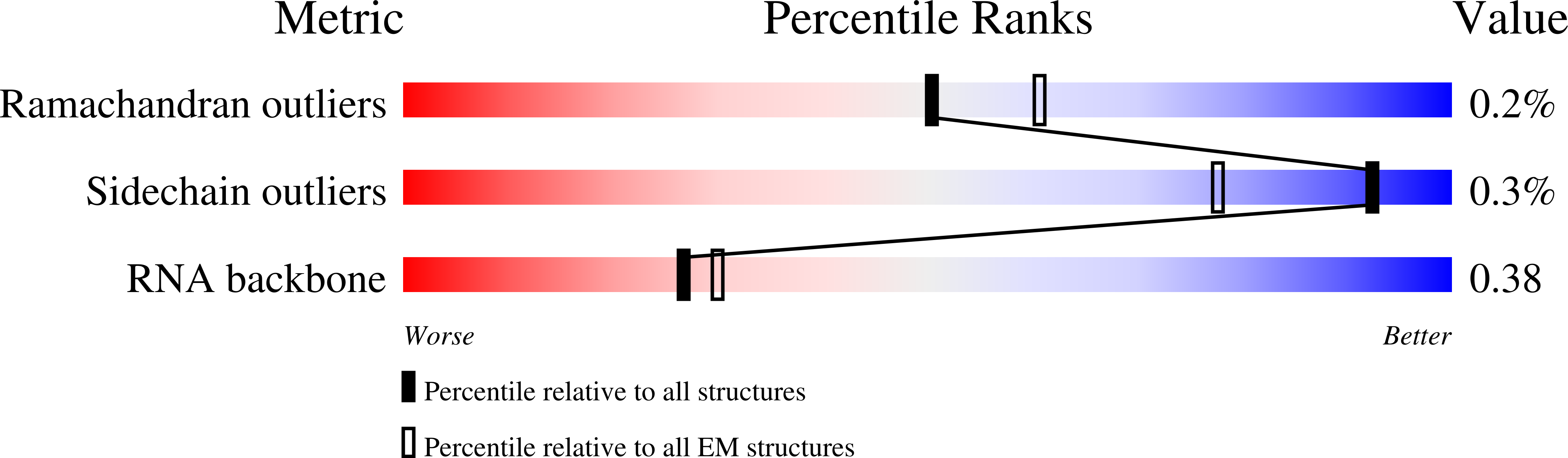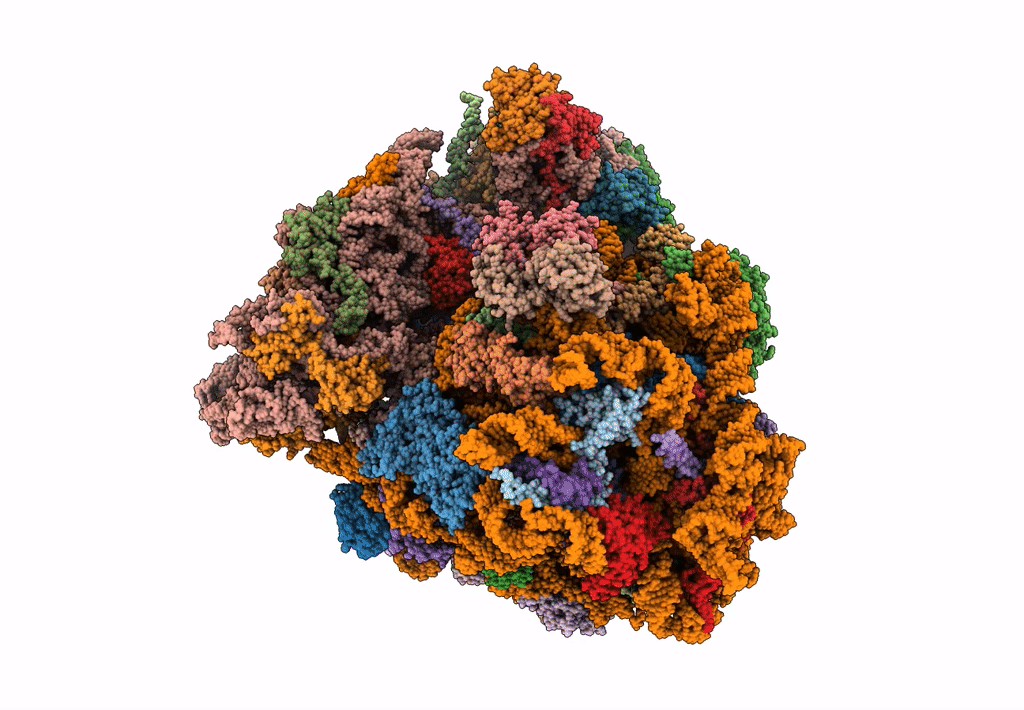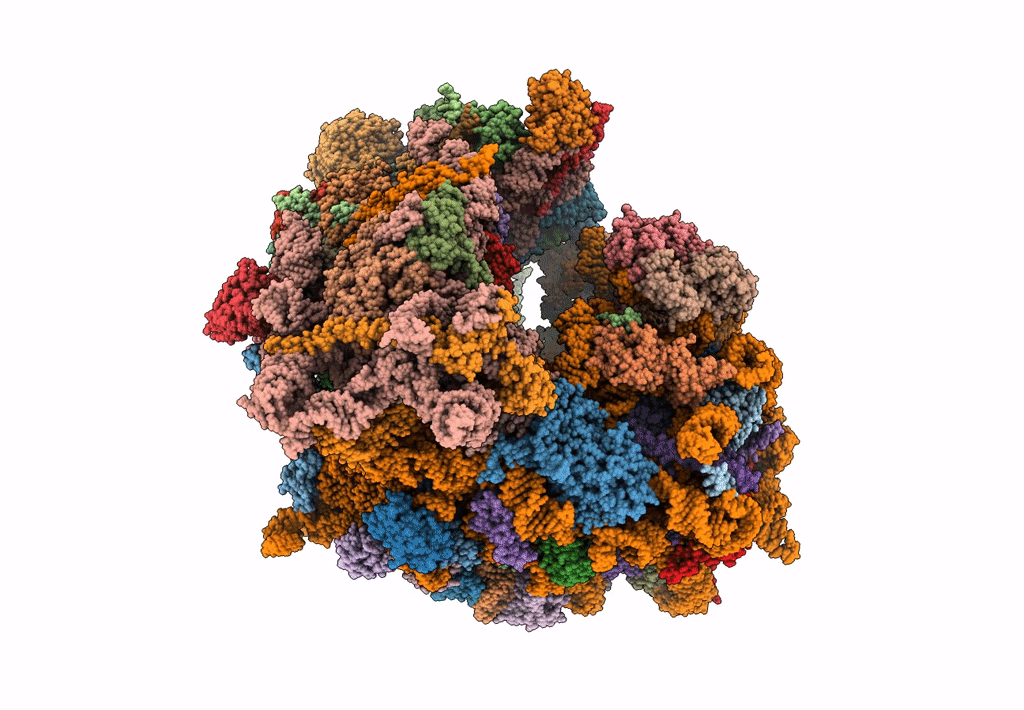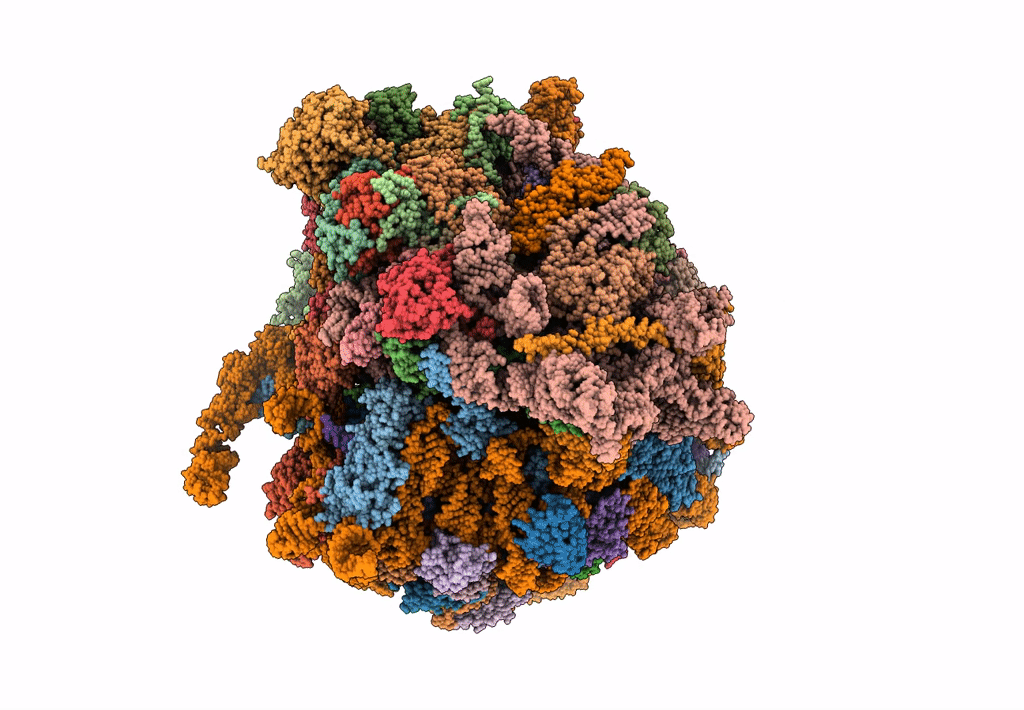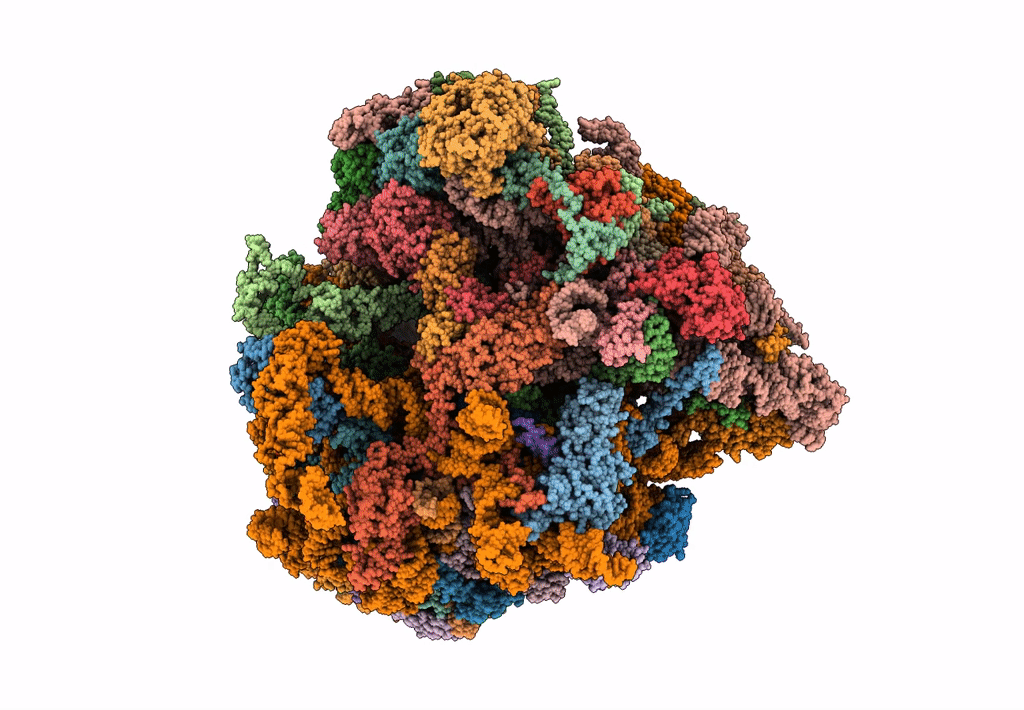
Deposition Date
2020-08-05
Release Date
2020-12-30
Last Version Date
2025-12-17
Entry Detail
PDB ID:
7A01
Keywords:
Title:
The Halastavi arva virus intergenic region IRES promotes translation by the simplest possible initiation mechanism
Biological Source:
Source Organism(s):
Halastavi arva RNA virus (Taxon ID: 1088890)
Oryctolagus cuniculus (Taxon ID: 9986)
Oryctolagus cuniculus (Taxon ID: 9986)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.60 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE