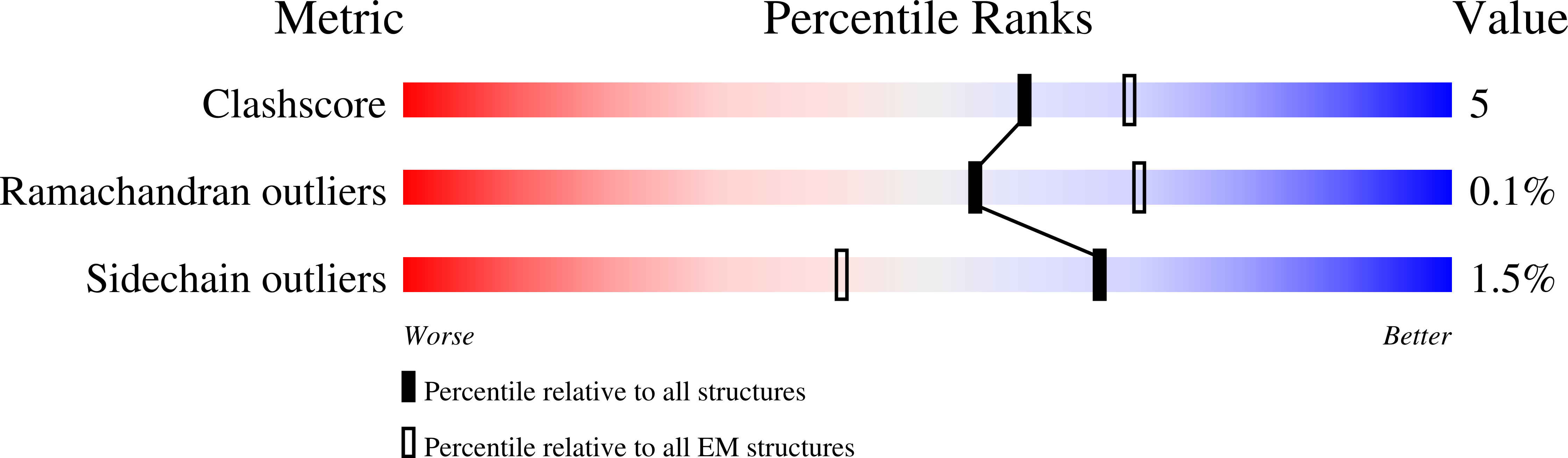
Deposition Date
2020-07-08
Release Date
2020-07-22
Last Version Date
2024-11-13
Entry Detail
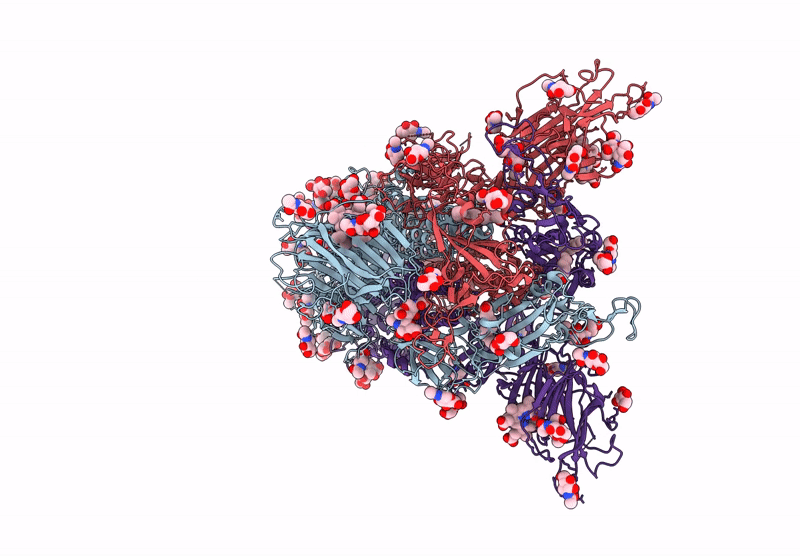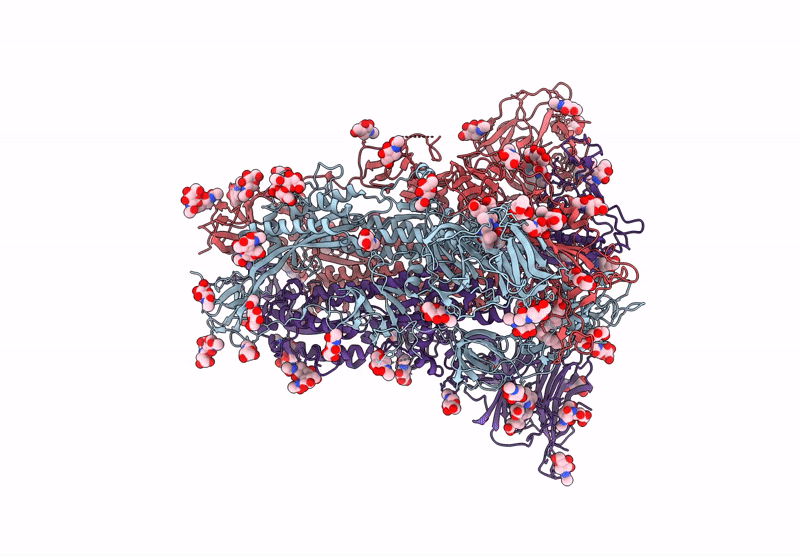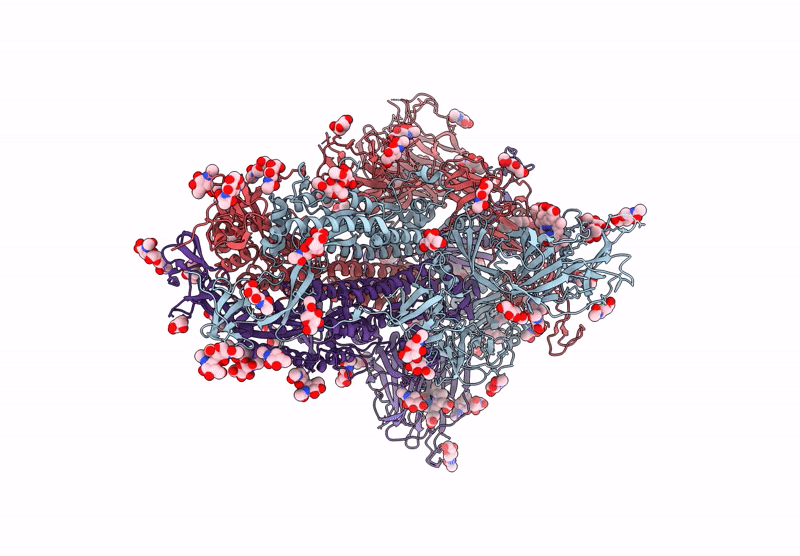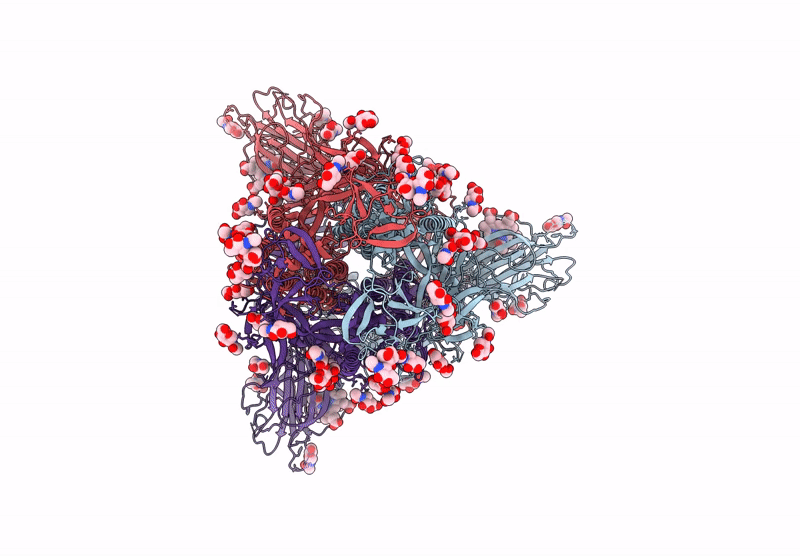
PDB ID:
6ZP2
Keywords:
Title:
Structure of SARS-CoV-2 Spike Protein Trimer (K986P, V987P, single Arg S1/S2 cleavage site) in Locked State
Biological Source:
Source Organism(s):
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.10 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE