Deposition Date
2020-07-07
Release Date
2021-06-16
Last Version Date
2024-01-31
Entry Detail
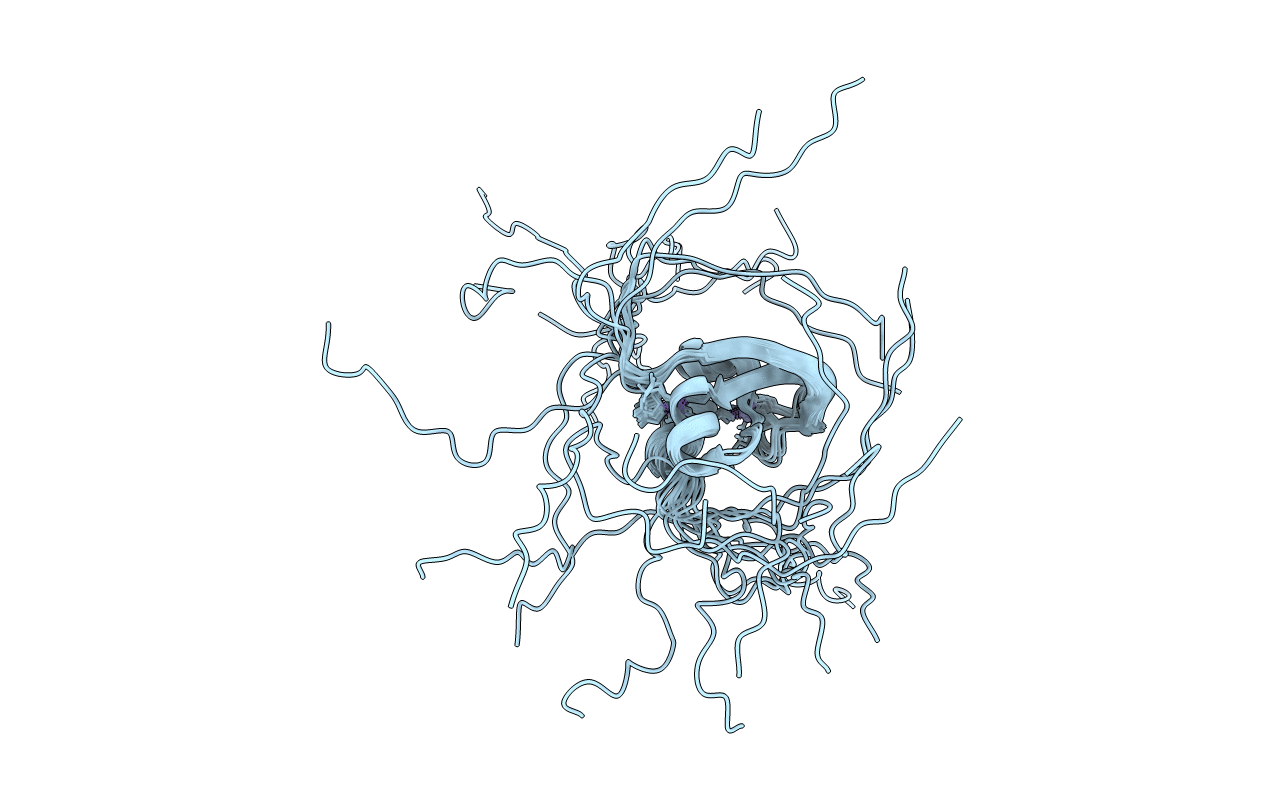
PDB ID:
6ZOP
Keywords:
Title:
Structure of the cysteine-rich domain of PiggyMac, a domesticated PiggyBac transposase involved in programmed genome rearrangements
Biological Source:
Source Organism(s):
Paramecium tetraurelia (Taxon ID: 5888)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
200
Conformers Submitted:
15
Selection Criteria:
target function


