Deposition Date
2020-07-07
Release Date
2020-12-23
Last Version Date
2024-01-31
Entry Detail
PDB ID:
6ZO1
Keywords:
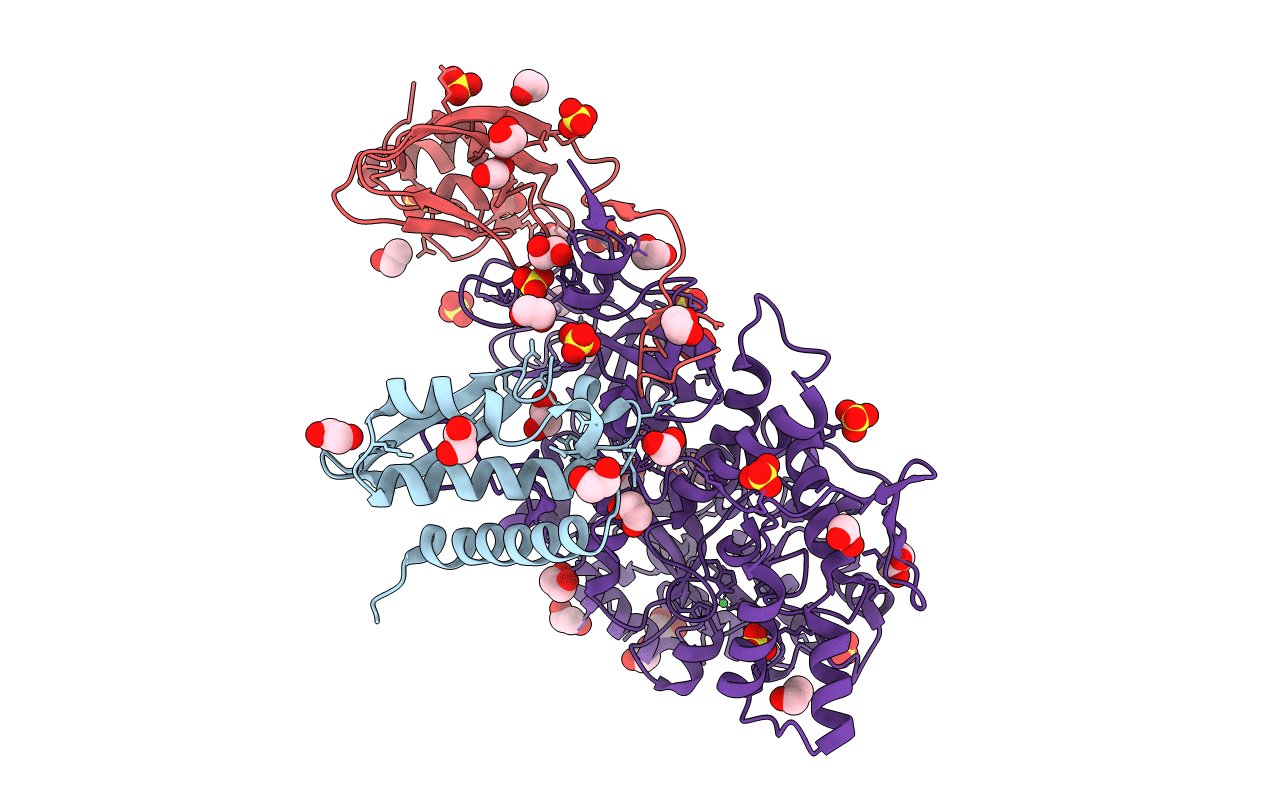
Title:
1.61 A resolution 3,5-dimethylcatechol (3,5-dimethylbenzene-1,2-diol) inhibited Sporosarcina pasteurii urease
Biological Source:
Source Organism:
Sporosarcina pasteurii (Taxon ID: 1474)
Method Details:
Experimental Method:
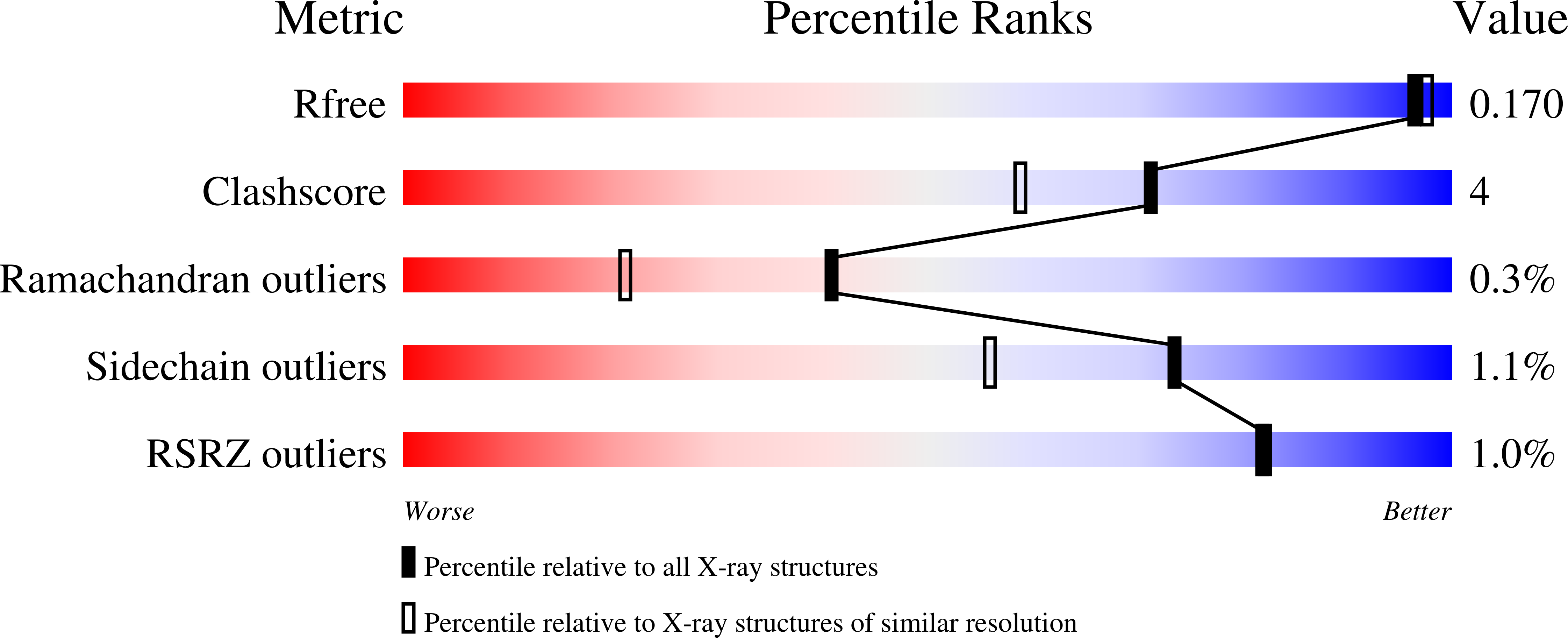
Resolution:
1.61 Å
R-Value Free:
0.15
R-Value Work:
0.13
Space Group:
P 63 2 2