Deposition Date
2020-06-11
Release Date
2020-07-01
Last Version Date
2024-01-24
Entry Detail
PDB ID:
6ZCO
Keywords:
Title:
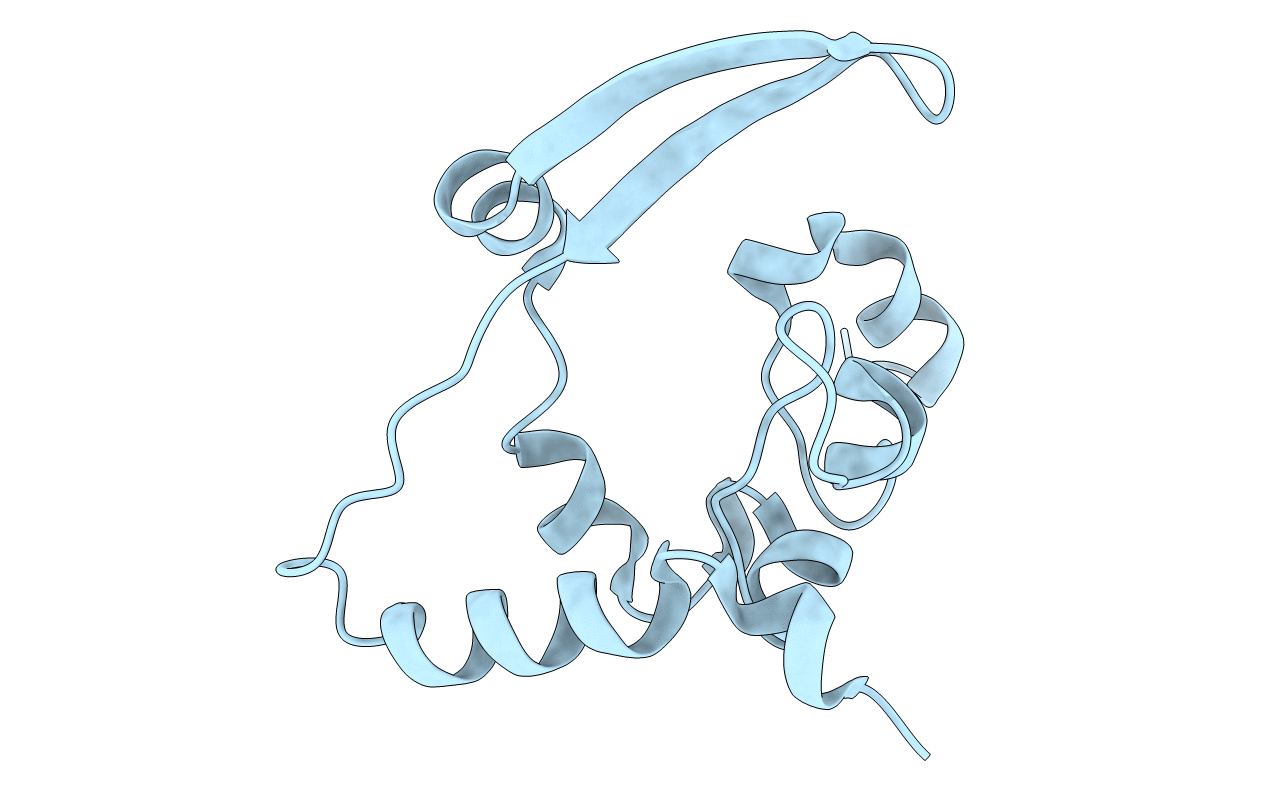
Crystal Structure of C-terminal Dimerization Domain of Nucleocapsid Phosphoprotein from SARS-CoV-2, crystal form II
Biological Source:
Source Organism:
Host Organism:
Method Details:
Experimental Method:
Resolution:
1.36 Å
R-Value Free:
0.19
R-Value Work:
0.14
R-Value Observed:
0.15
Space Group:
I 41


