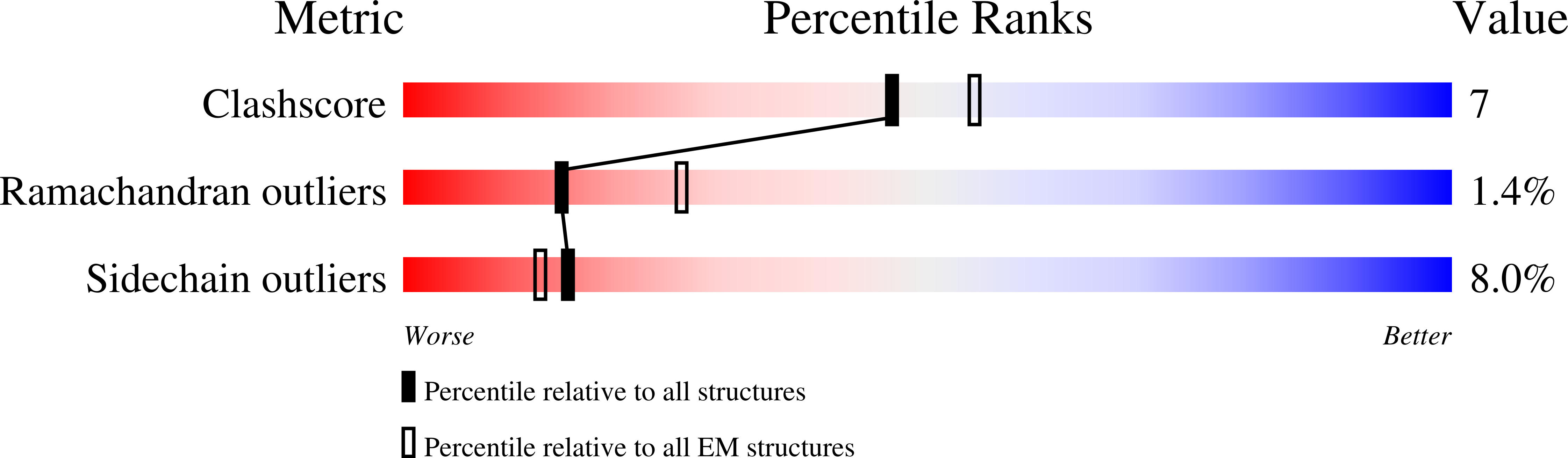
Deposition Date
2020-06-01
Release Date
2020-07-15
Last Version Date
2024-05-15
Entry Detail
PDB ID:
6Z7P
Keywords:
Title:
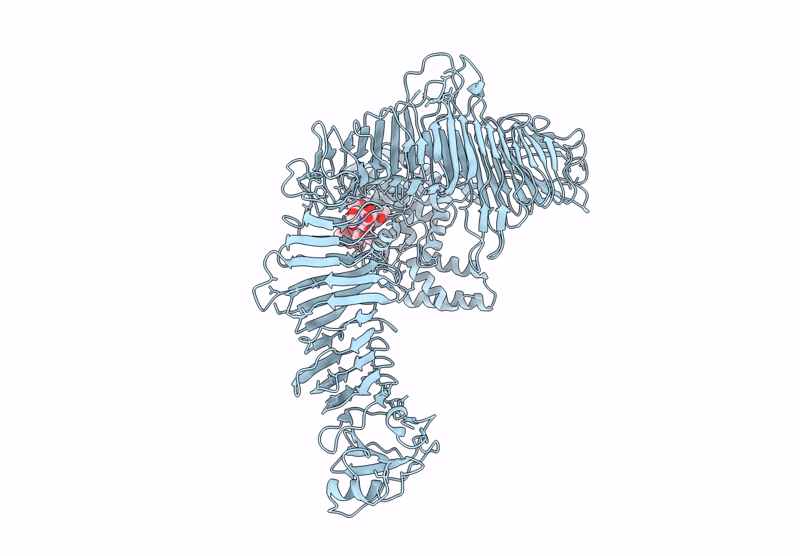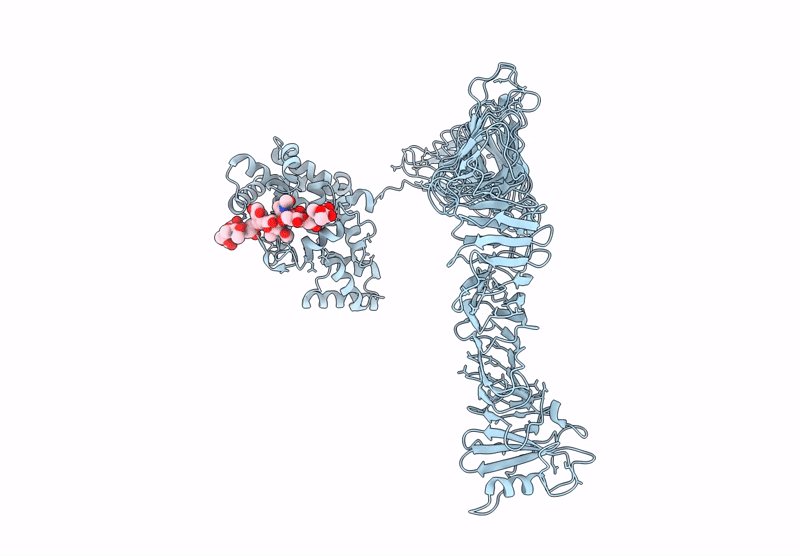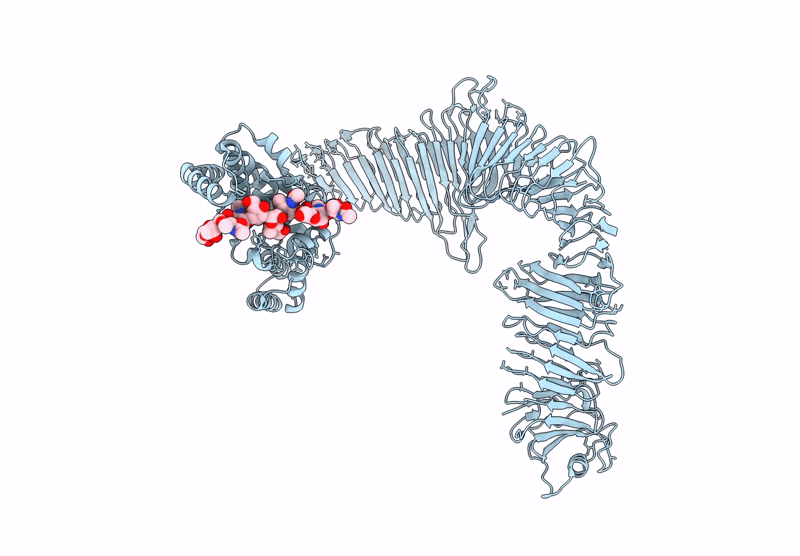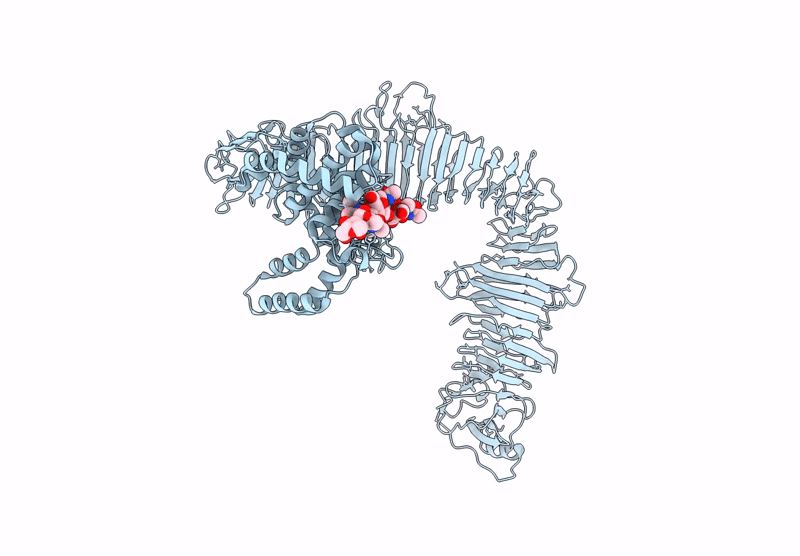
Composite model of the Caulobacter crescentus S-layer bound to the O-antigen of lipopolysaccharide
Biological Source:
Source Organism(s):
Method Details:
Experimental Method:
Resolution:
4.80 Å
Aggregation State:
CELL
Reconstruction Method:
SUBTOMOGRAM AVERAGING