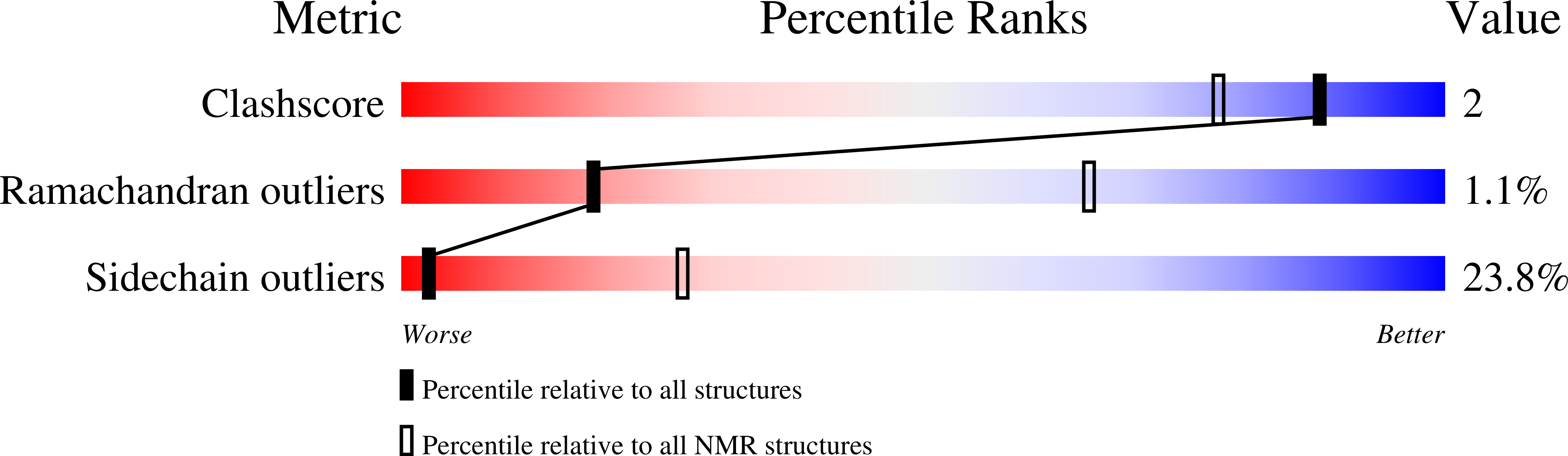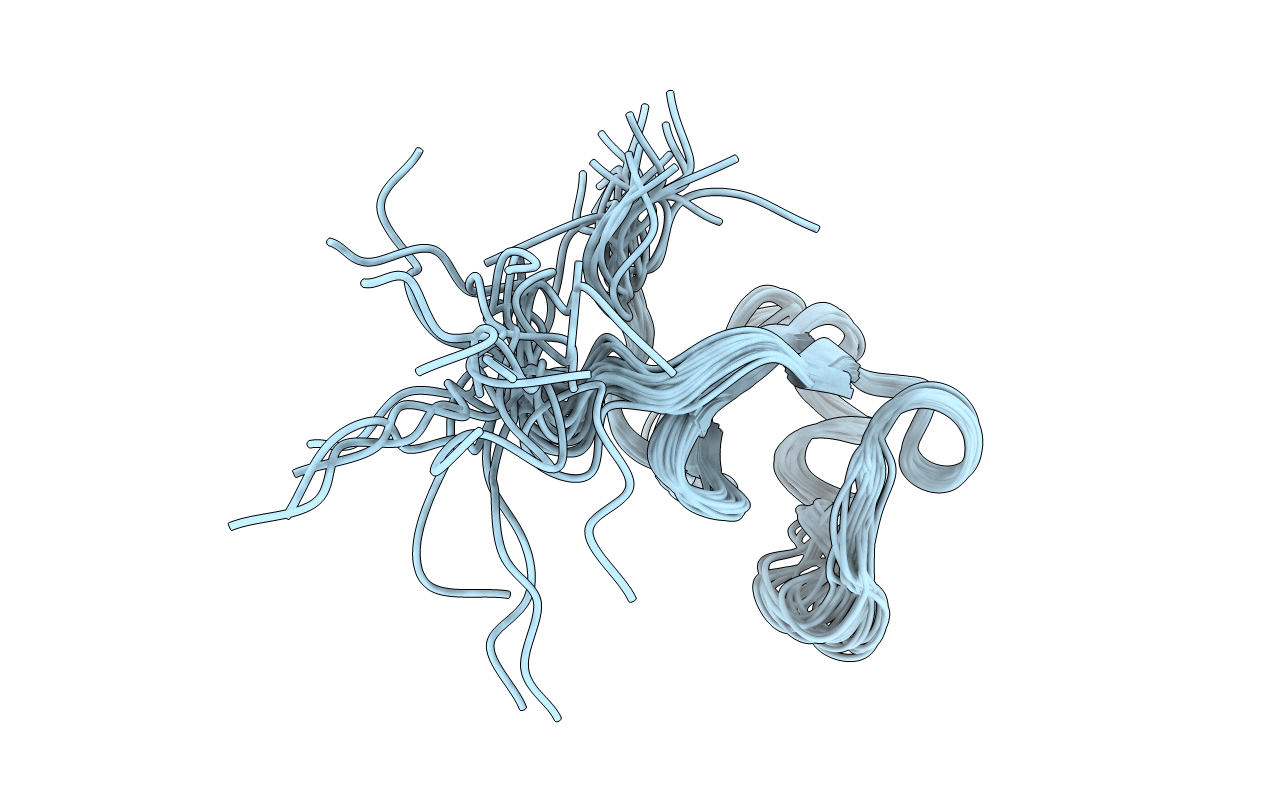
Deposition Date
2020-05-22
Release Date
2021-05-05
Last Version Date
2024-11-20
Entry Detail
PDB ID:
6Z41
Keywords:
Title:
NMR solution structure of the carbohydrate-binding module family 73 (CBM73) from Cellvibrio japonicus CjLPMO10A
Biological Source:
Source Organism(s):
Cellvibrio japonicus Ueda107 (Taxon ID: 498211)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
256
Conformers Submitted:
20
Selection Criteria:
target function