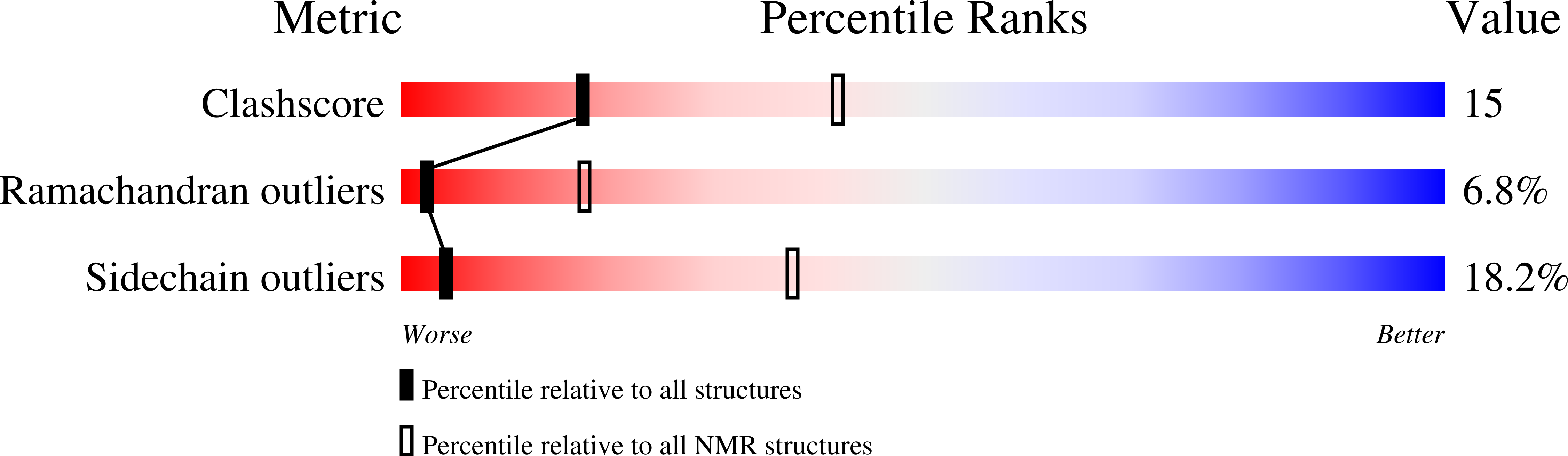
Deposition Date
2020-05-15
Release Date
2021-05-26
Last Version Date
2024-06-19
Entry Detail
PDB ID:
6Z29
Keywords:
Title:
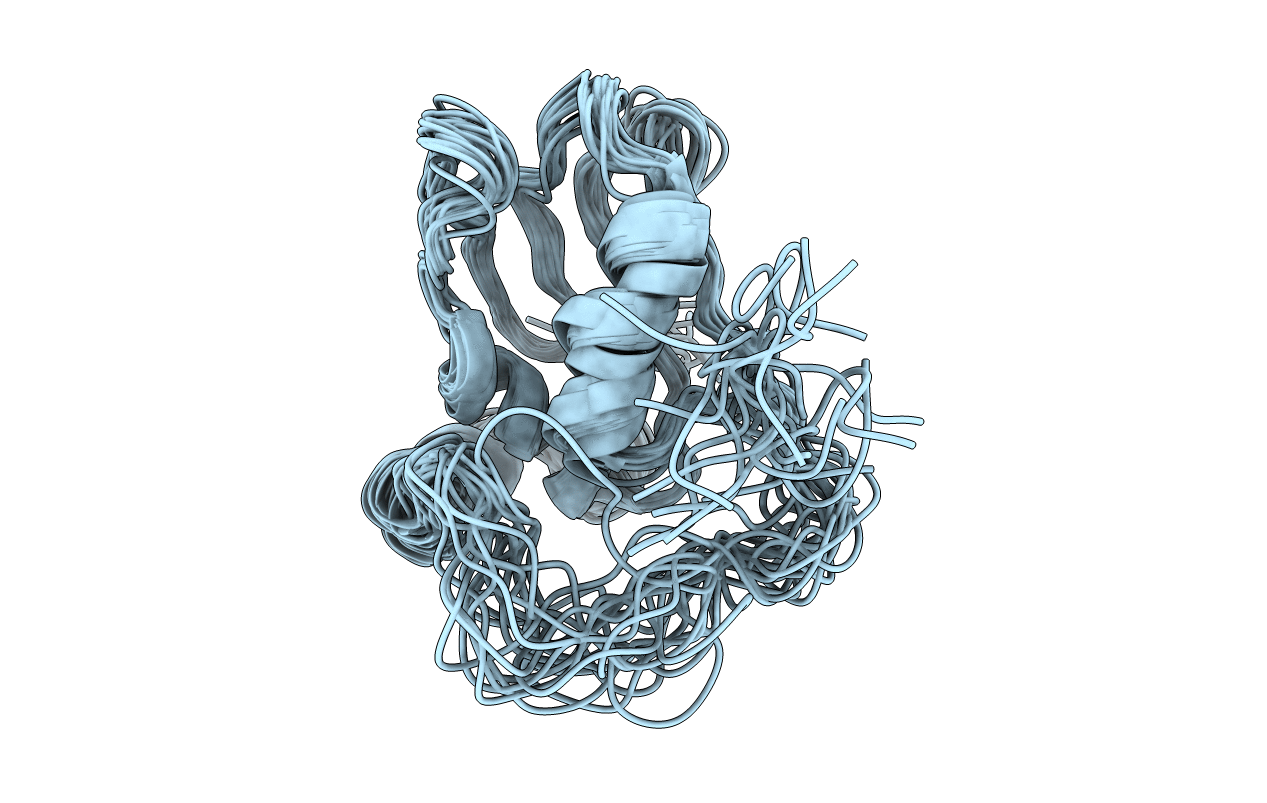
Structure of eIF4G1 (37-49) - PUB1 RRM3 chimera in solution
Biological Source:
Source Organism(s):
Saccharomyces cerevisiae S288C (Taxon ID: 559292)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
20
Selection Criteria:
target function