Deposition Date
2020-02-14
Release Date
2020-06-17
Last Version Date
2024-05-15
Entry Detail
PDB ID:
6Y1X
Keywords:
Title:
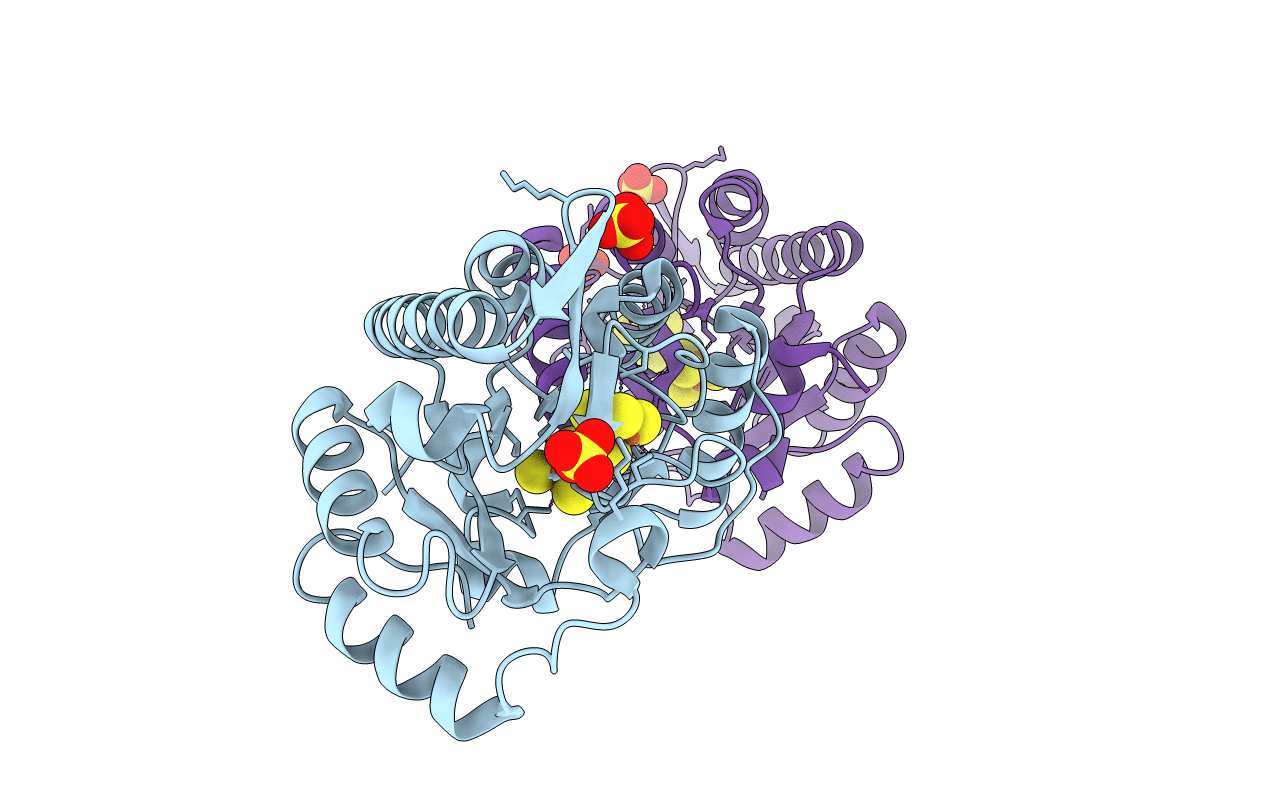
X-ray structure of the radical SAM protein NifB, a key nitrogenase maturating enzyme
Biological Source:
Source Organism:
Host Organism:
Method Details:
Experimental Method:
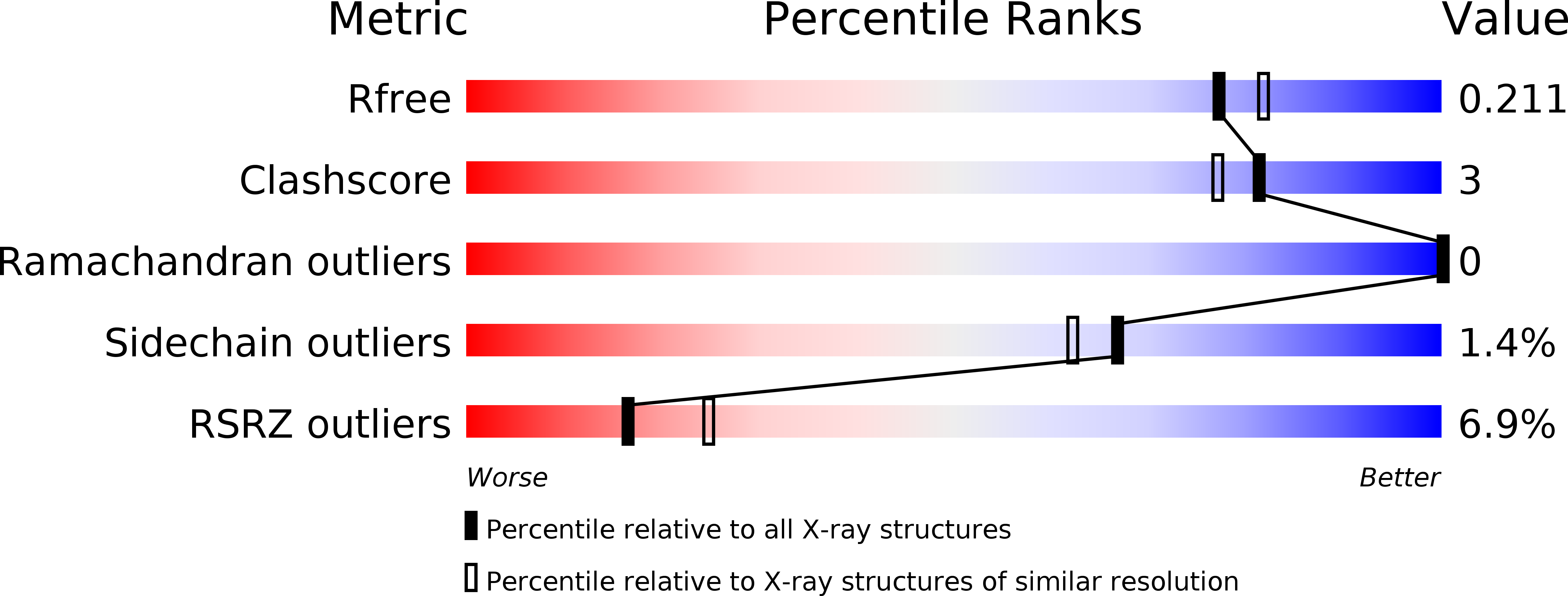
Resolution:
1.95 Å
R-Value Free:
0.21
R-Value Work:
0.17
R-Value Observed:
0.17
Space Group:
P 1