Deposition Date
2020-06-02
Release Date
2021-02-17
Last Version Date
2023-10-18
Entry Detail
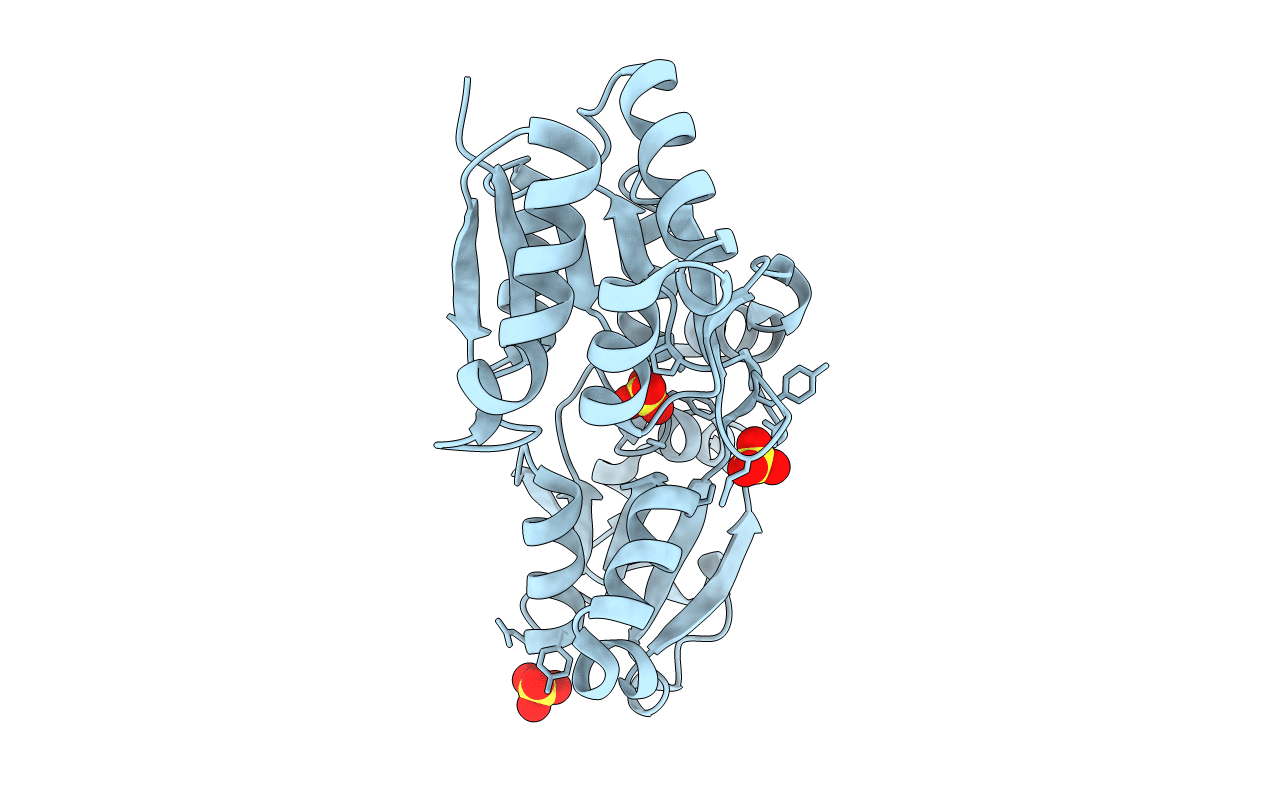
PDB ID:
6X8W
Keywords:
Title:
Structure of ArrX Y138A mutant protein bound to sulfate from Chrysiogenes arsenatis
Biological Source:
Source Organism(s):
Chrysiogenes arsenatis (Taxon ID: 309797)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
1.97 Å
R-Value Free:
0.20
R-Value Work:
0.17
Space Group:
P 21 21 21


