Deposition Date
2020-05-22
Release Date
2020-06-24
Last Version Date
2023-10-18
Entry Detail
PDB ID:
6X4P
Keywords:
Title:
Human cyclophilin A bound to a series of acylcic and macrocyclic inhibitors: (2R,5S,11S,14S,18E)-2,11,17,17-tetramethyl-14-(propan-2-yl)-15-oxa-3,9,12,26,29-pentaazatetracyclo[18.5.3.1~5,9~.0~23,27~]nonacosa-1(25),18,20(28),21,23,26-hexaene-4,10,13,16-tetrone (compound 28)
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
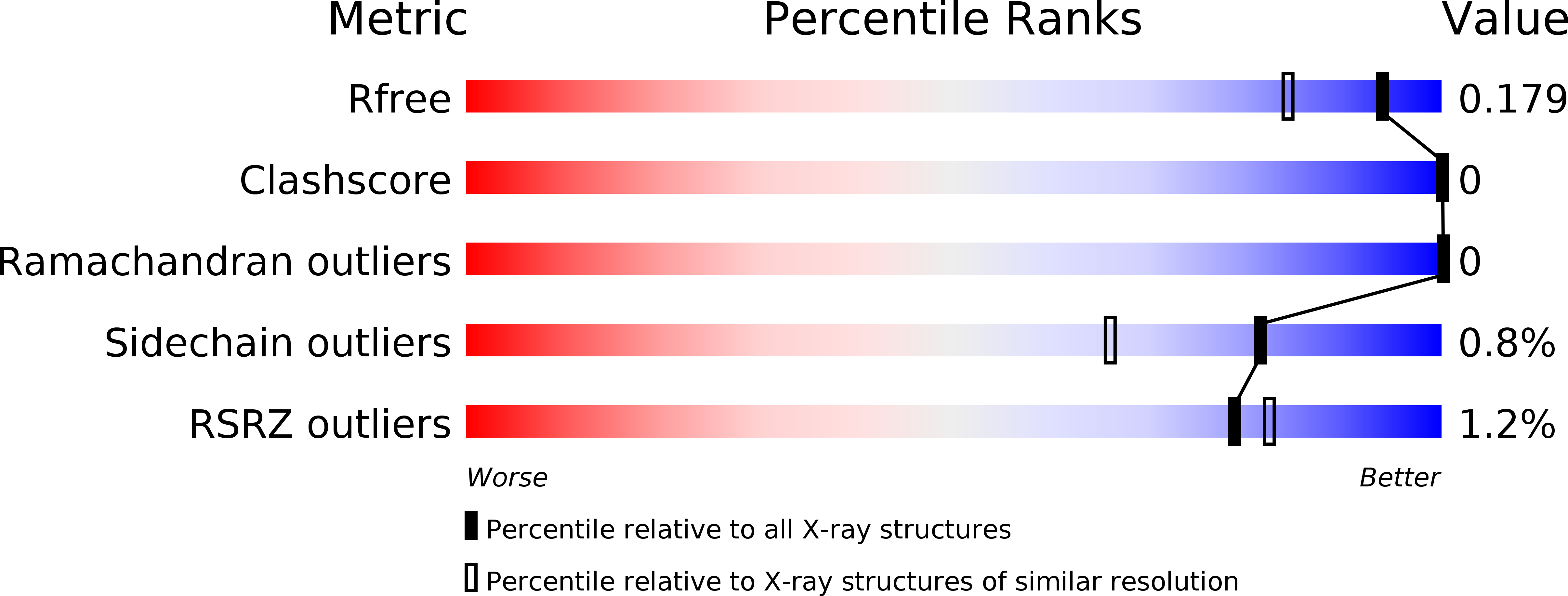
Resolution:
1.50 Å
R-Value Free:
0.17
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
P 21 21 21