Deposition Date
2020-05-10
Release Date
2021-01-20
Last Version Date
2024-11-13
Entry Detail
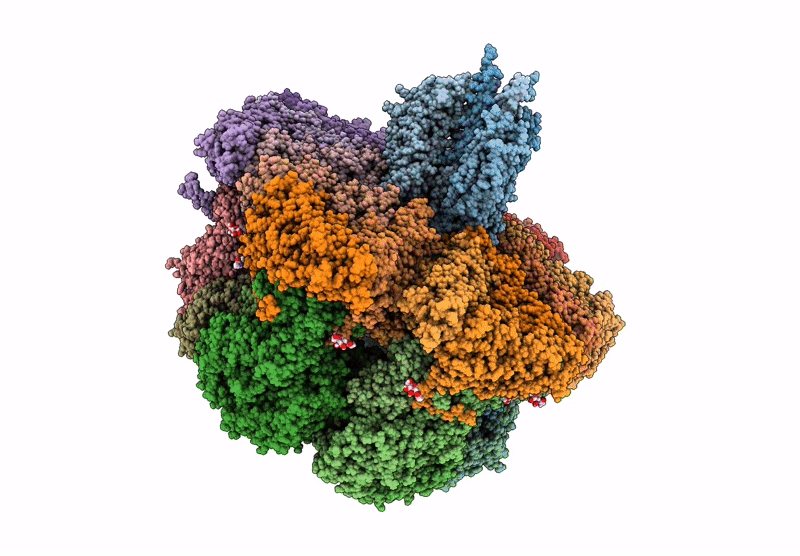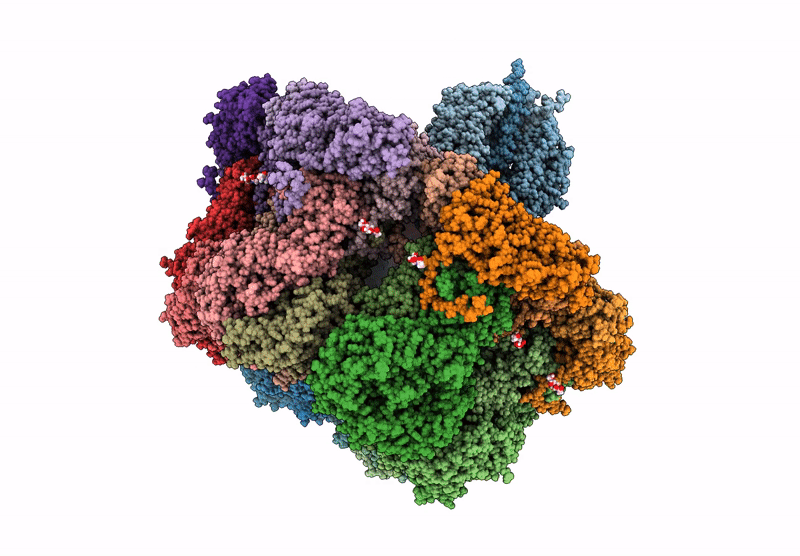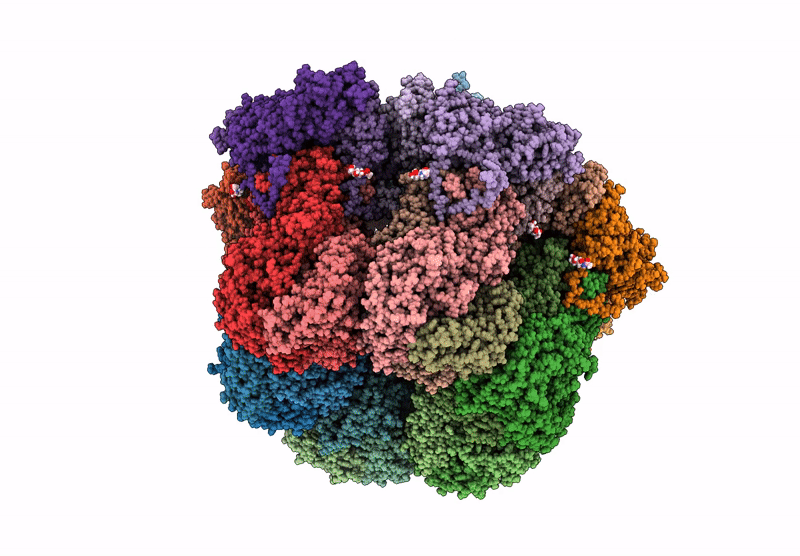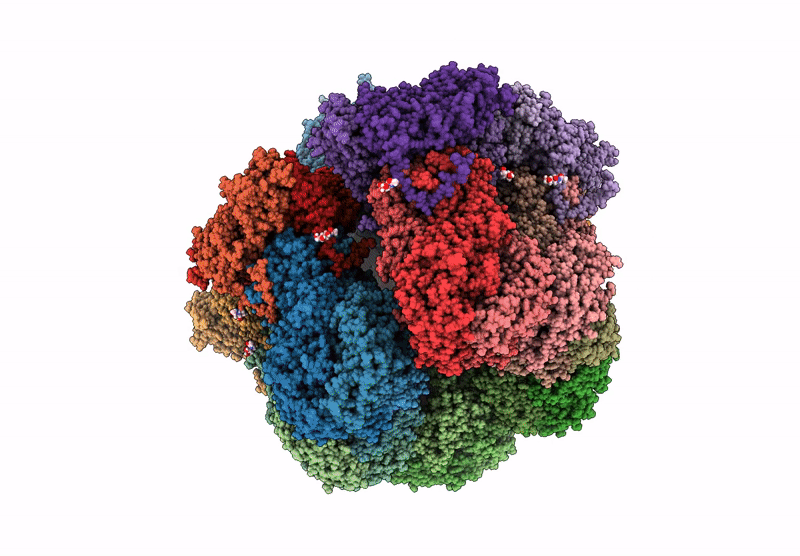
PDB ID:
6WXG
Keywords:
Title:
Cryo-EM reconstruction of VP5*/VP8* assembly from rhesus rotavirus particles - Reversed conformation
Biological Source:
Source Organism(s):
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.30 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE