Deposition Date
2020-05-06
Release Date
2020-11-18
Last Version Date
2024-05-29
Entry Detail
PDB ID:
6WVJ
Keywords:
Title:
Cryo-EM structure of Bacillus subtilis RNA Polymerase elongation complex
Biological Source:
Source Organism:
Bacillus subtilis (strain 168) (Taxon ID: 224308)
Bacillus subtilis (Taxon ID: 1423)
Escherichia coli BL21(DE3) (Taxon ID: 469008)
Bacillus subtilis (Taxon ID: 1423)
Escherichia coli BL21(DE3) (Taxon ID: 469008)
Host Organism:
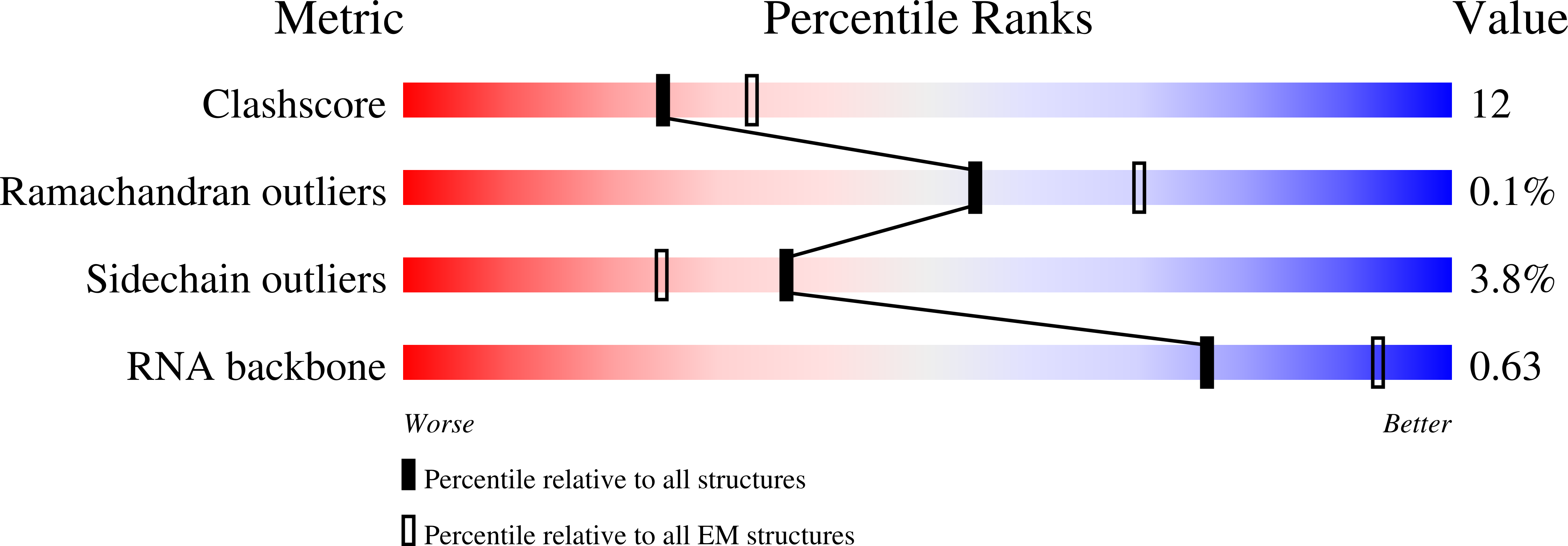
Method Details:
Experimental Method:
Resolution:
3.36 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE