Deposition Date
2020-04-30
Release Date
2020-08-05
Last Version Date
2024-10-23
Entry Detail
PDB ID:
6WS1
Keywords:
Title:
Crystal structure of human phenylethanolamine N-methyltransferase (PNMT) in complex with (2S)-2-amino-4-((((2R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)(3-(7,8-dichloro-1,2,3,4-tetrahydroisoquinolin-4-yl)propyl)amino)butanoic acid and AdoHcy (SAH)
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
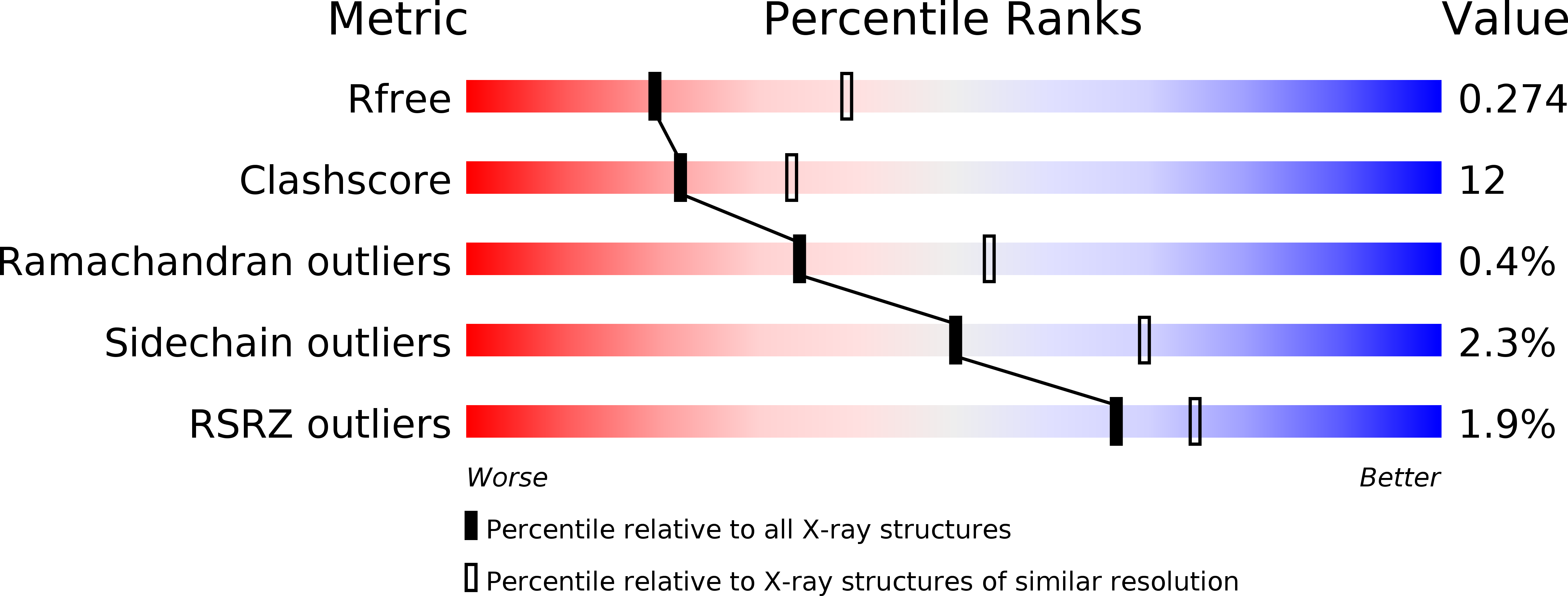
Resolution:
2.76 Å
R-Value Free:
0.27
R-Value Work:
0.22
R-Value Observed:
0.22
Space Group:
P 43 21 2