Deposition Date
2020-04-16
Release Date
2021-05-19
Last Version Date
2023-10-18
Entry Detail
PDB ID:
6WKO
Keywords:
Title:
Structure of an influenza C virus hemagglutinin-esterase-fusion (HEF2) intermediate
Biological Source:
Source Organism:
Host Organism:
Method Details:
Experimental Method:
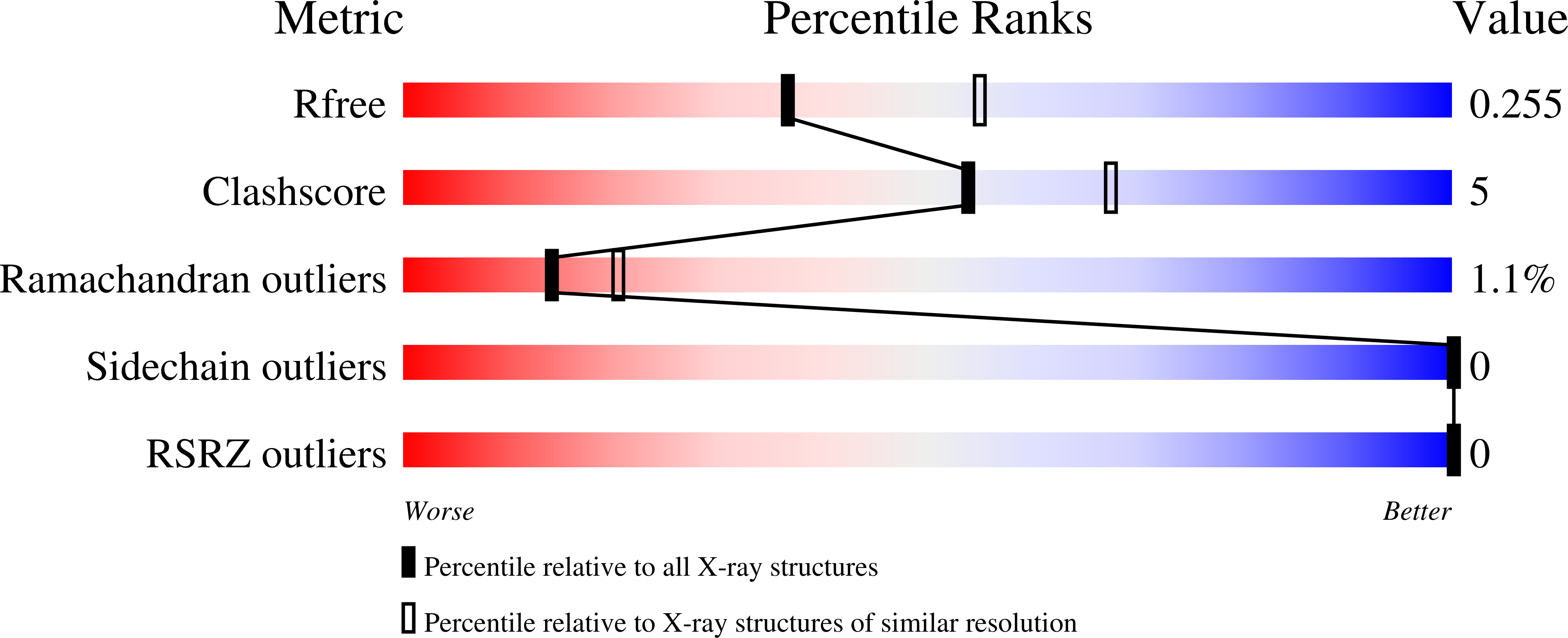
Resolution:
2.40 Å
R-Value Free:
0.25
R-Value Work:
0.19
R-Value Observed:
0.20
Space Group:
P 21 3