Deposition Date
2020-04-10
Release Date
2021-04-14
Last Version Date
2023-10-25
Entry Detail
PDB ID:
6WIJ
Keywords:
Title:
The crystal structure of the 2009/H1N1/California PA endonuclease mutant I38T in complex with SJ000986448
Biological Source:
Source Organism(s):
Influenza A virus (Taxon ID: 11320)
Influenza A virus (A/Luxembourg/43/2009(H1N1)) (Taxon ID: 655278)
Influenza A virus (A/Luxembourg/43/2009(H1N1)) (Taxon ID: 655278)
Expression System(s):
Method Details:
Experimental Method:
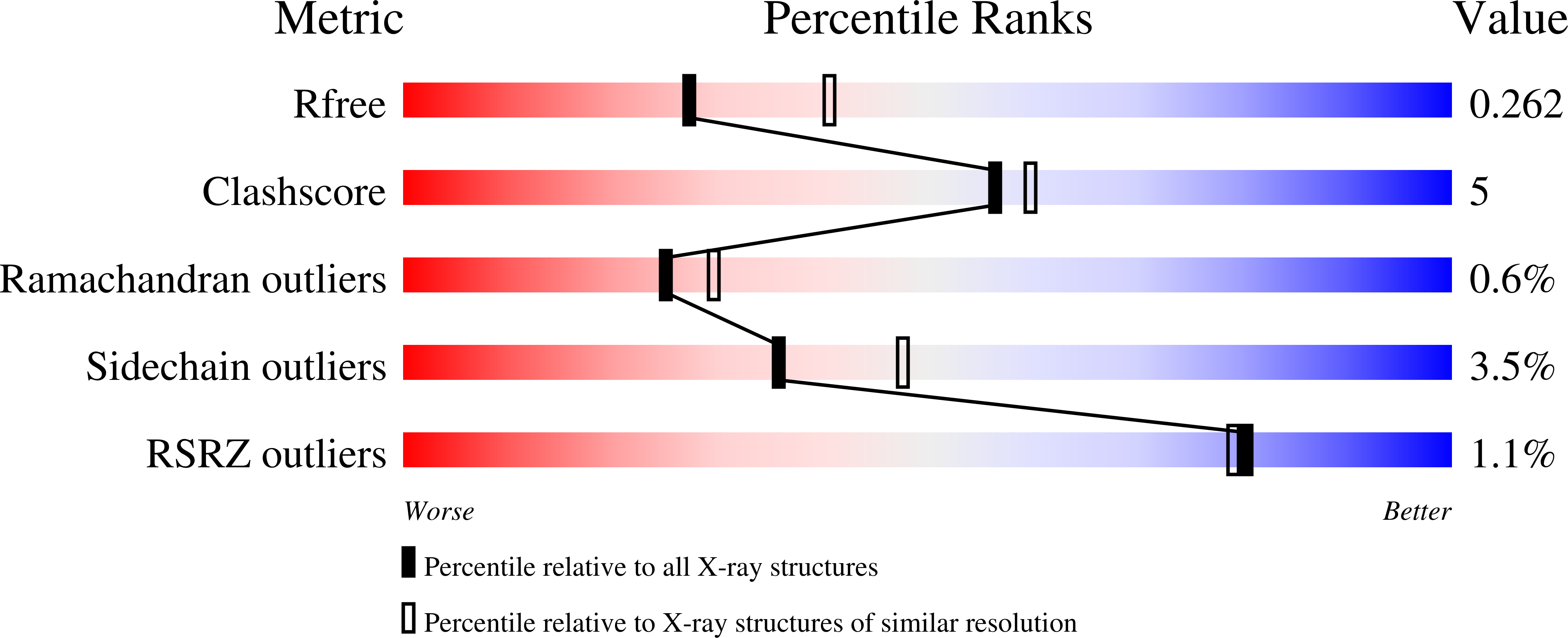
Resolution:
2.44 Å
R-Value Free:
0.26
R-Value Work:
0.21
R-Value Observed:
0.21
Space Group:
I 4 2 2