Deposition Date
2020-03-28
Release Date
2020-11-18
Last Version Date
2023-10-18
Entry Detail
PDB ID:
6WC0
Keywords:
Title:
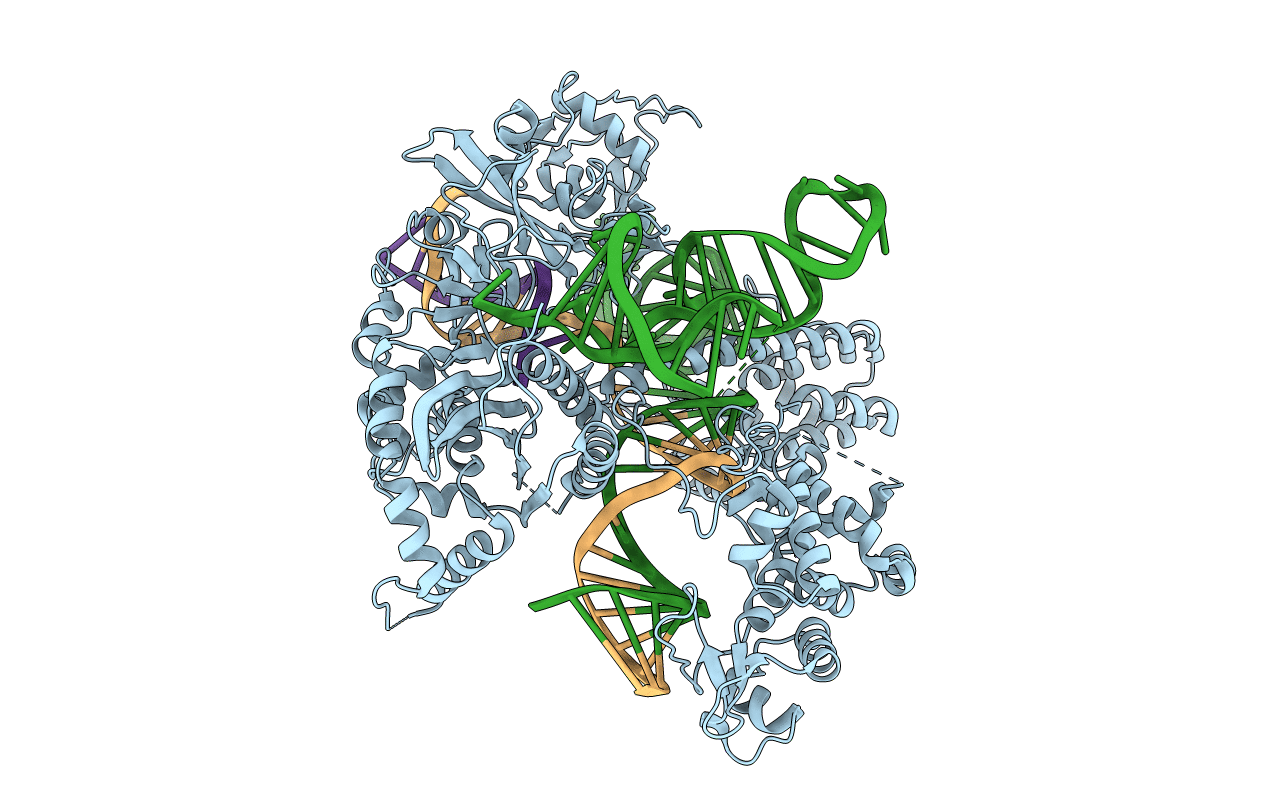
Crystal structure of AceCas9 bound with guide RNA and DNA with 5'-NNNTC-3' PAM
Biological Source:
Source Organism(s):
Acidothermus cellulolyticus (strain ATCC 43068 / 11B) (Taxon ID: 351607)
synthetic construct (Taxon ID: 32630)
synthetic construct (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.61 Å
R-Value Free:
0.30
R-Value Work:
0.23
R-Value Observed:
0.24
Space Group:
P 21 21 21


