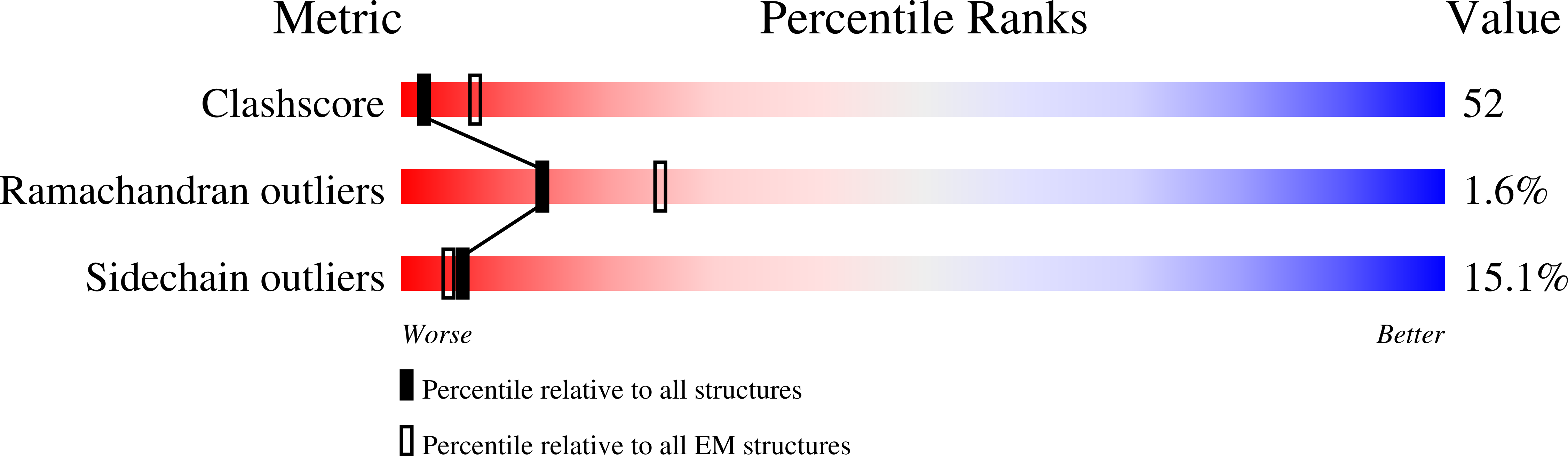
Deposition Date
2020-03-17
Release Date
2020-06-24
Last Version Date
2024-11-13
Entry Detail
PDB ID:
6W6O
Keywords:
Title:
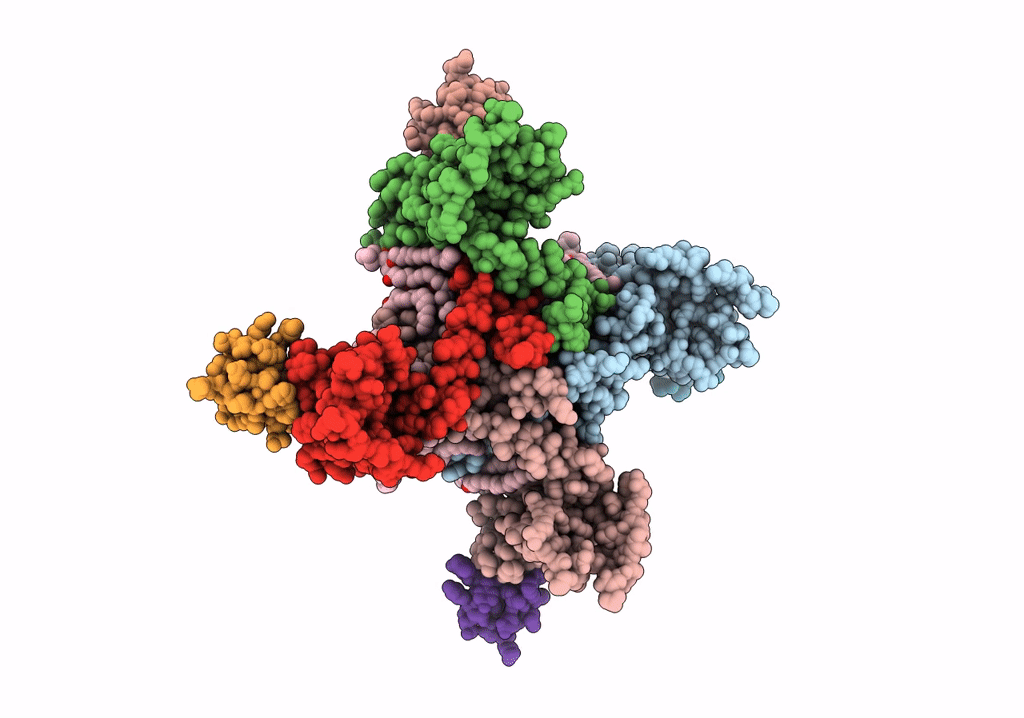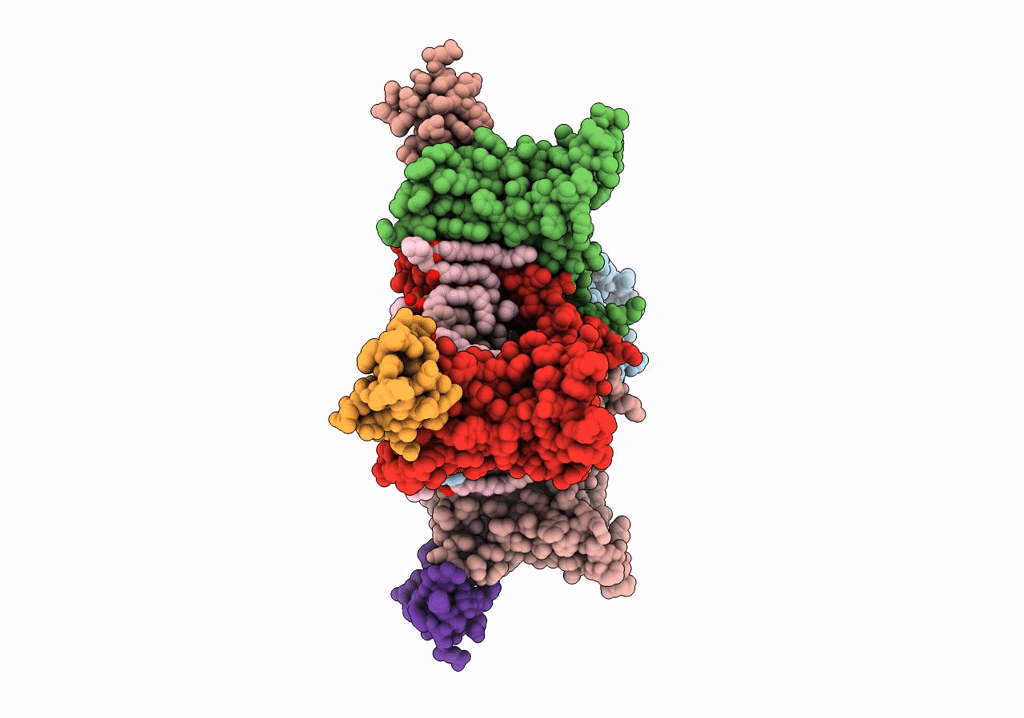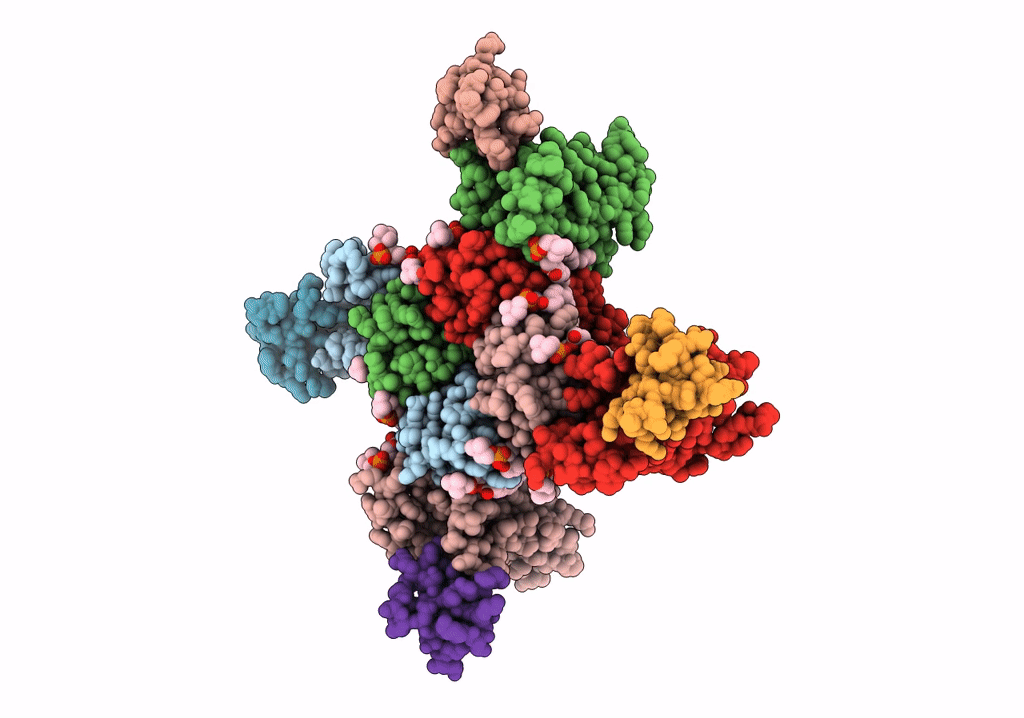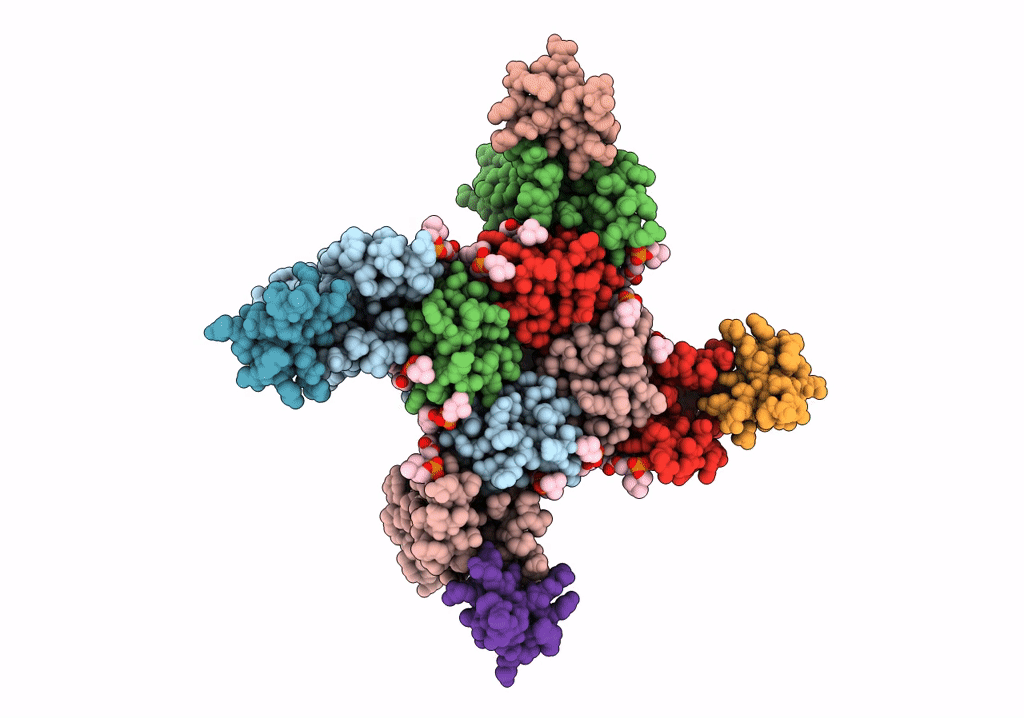
NaChBac-Nav1.7VSDII chimera and HWTX-IV complex
Biological Source:
Source Organism(s):
Bacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) (Taxon ID: 272558)
Homo sapiens (Taxon ID: 9606)
Cyriopagopus schmidti (Taxon ID: 29017)
Homo sapiens (Taxon ID: 9606)
Cyriopagopus schmidti (Taxon ID: 29017)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.20 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE