Deposition Date
2020-03-11
Release Date
2020-08-05
Last Version Date
2023-11-15
Entry Detail
PDB ID:
6W4M
Keywords:
Title:
CRYSTAL STRUCTURE OF THE ADCC-POTENT, WEAKLY NEUTRALIZING HIV ENV CO-RECEPTOR BINDING SITE ANTIBODY N12-I2 FAB IN COMPLEX WITH HIV-1 CLADE A/E GP120 AND M48U1
Biological Source:
Source Organism(s):
Human immunodeficiency virus 1 (Taxon ID: 11676)
Homo sapiens (Taxon ID: 9606)
synthetic construct (Taxon ID: 32630)
Homo sapiens (Taxon ID: 9606)
synthetic construct (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
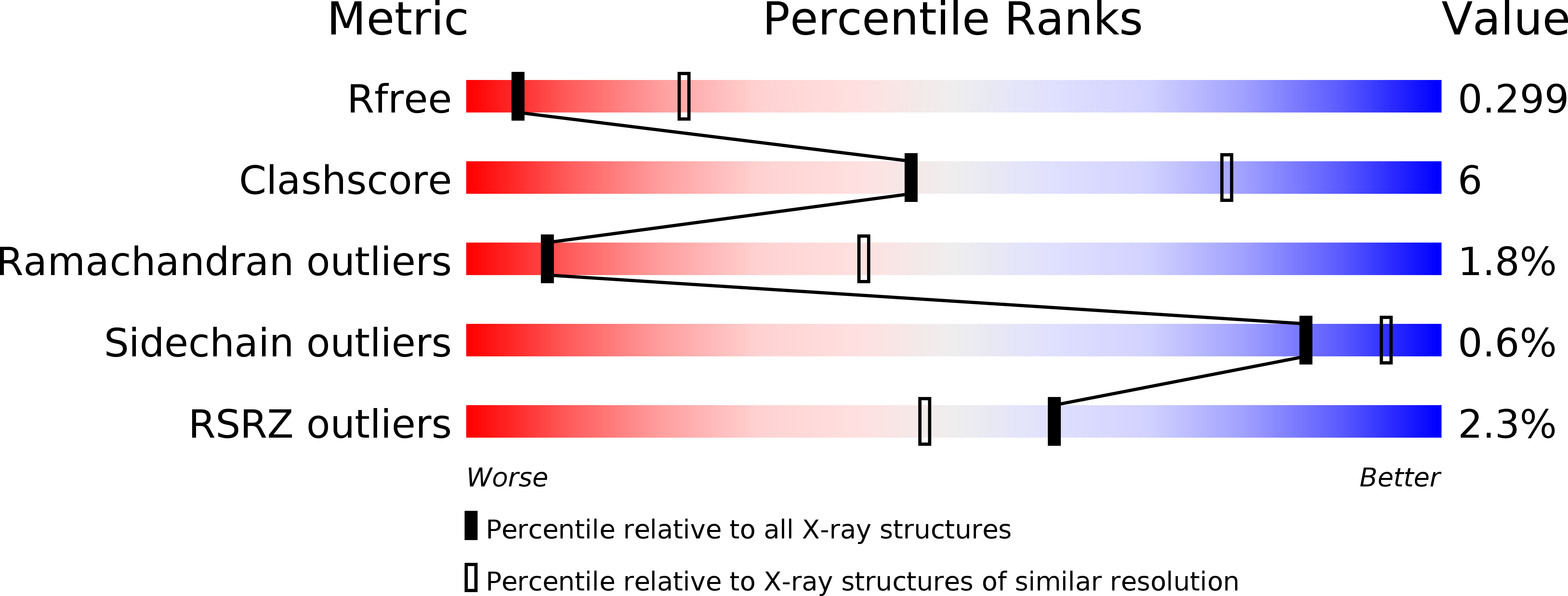
Resolution:
3.20 Å
R-Value Free:
0.30
R-Value Work:
0.24
R-Value Observed:
0.24
Space Group:
P 21 21 21