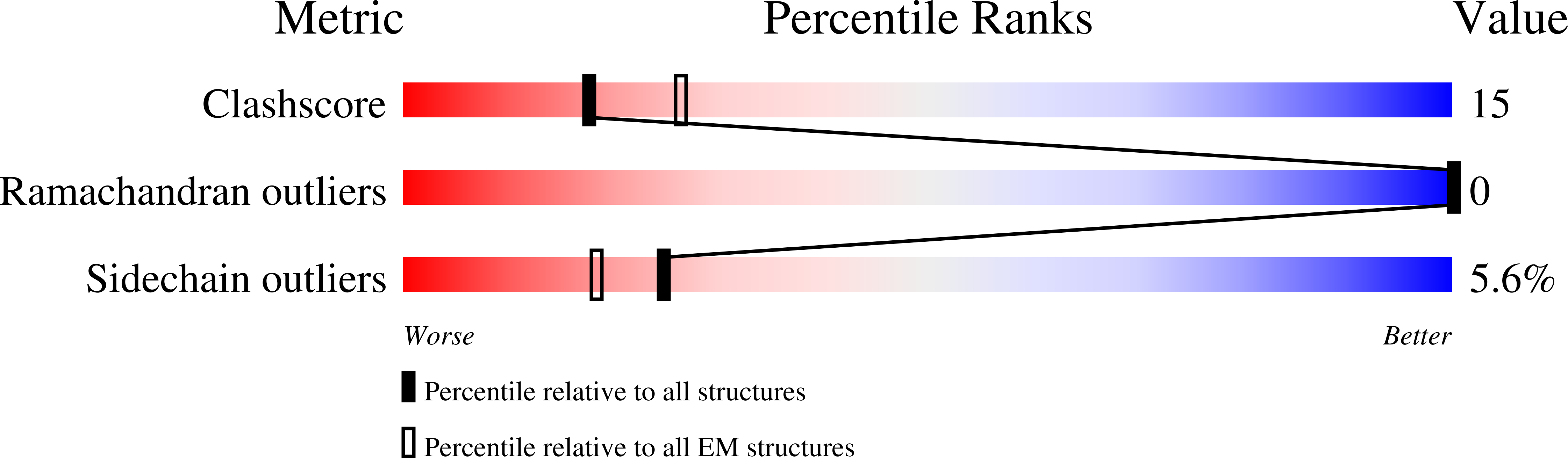Deposition Date
2020-02-22
Release Date
2020-04-29
Last Version Date
2024-11-13
Entry Detail
PDB ID:
6VXK
Keywords:
Title:
Cryo-EM Structure of the full-length A39R/PlexinC1 complex
Biological Source:
Source Organism(s):
Ectromelia virus (strain Moscow) (Taxon ID: 265874)
Homo sapiens (Taxon ID: 9606)
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.10 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE