Deposition Date
2020-02-13
Release Date
2020-02-26
Last Version Date
2023-10-11
Entry Detail
PDB ID:
6VTZ
Keywords:
Title:
Structure of a thiolation-reductase di-domain from an archaeal non-ribosomal peptide synthetase
Biological Source:
Source Organism:
Host Organism:
Method Details:
Experimental Method:
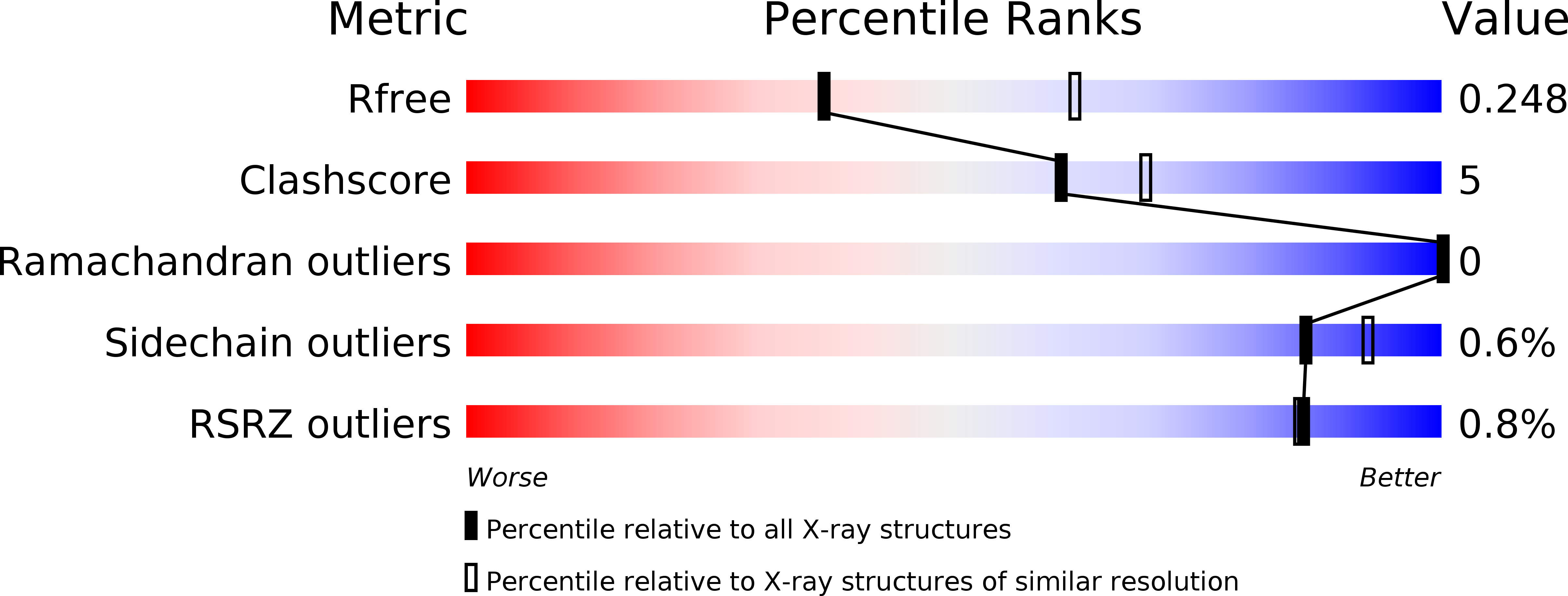
Resolution:
2.65 Å
R-Value Free:
0.24
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
P 32 2 1