Deposition Date
2020-02-03
Release Date
2020-07-29
Last Version Date
2024-03-06
Entry Detail
PDB ID:
6VPC
Keywords:
Title:
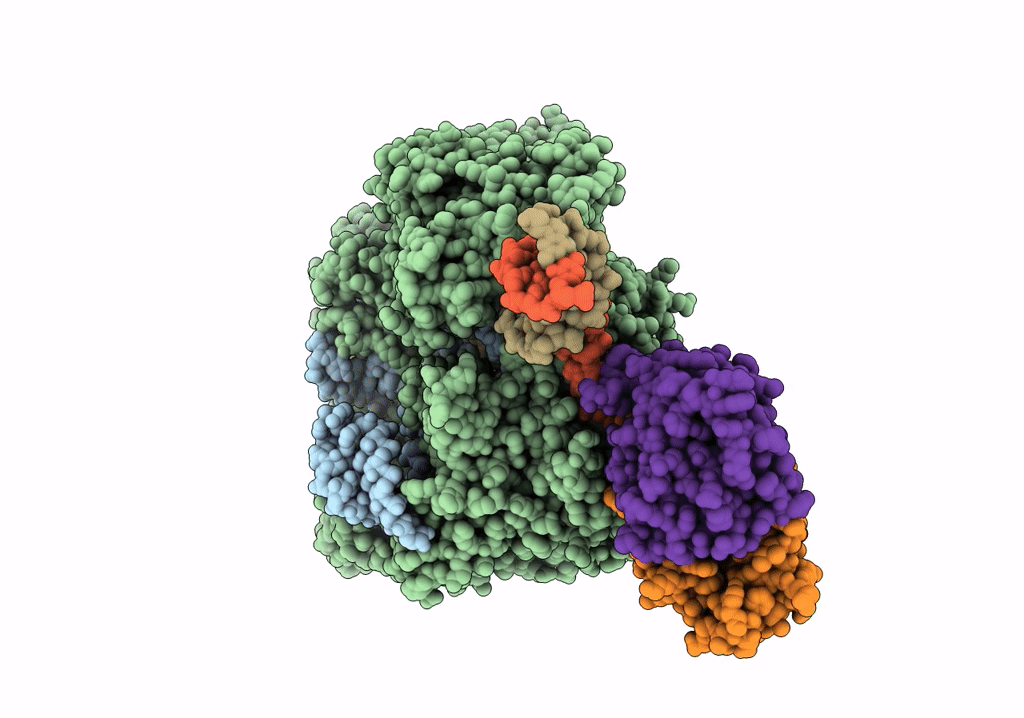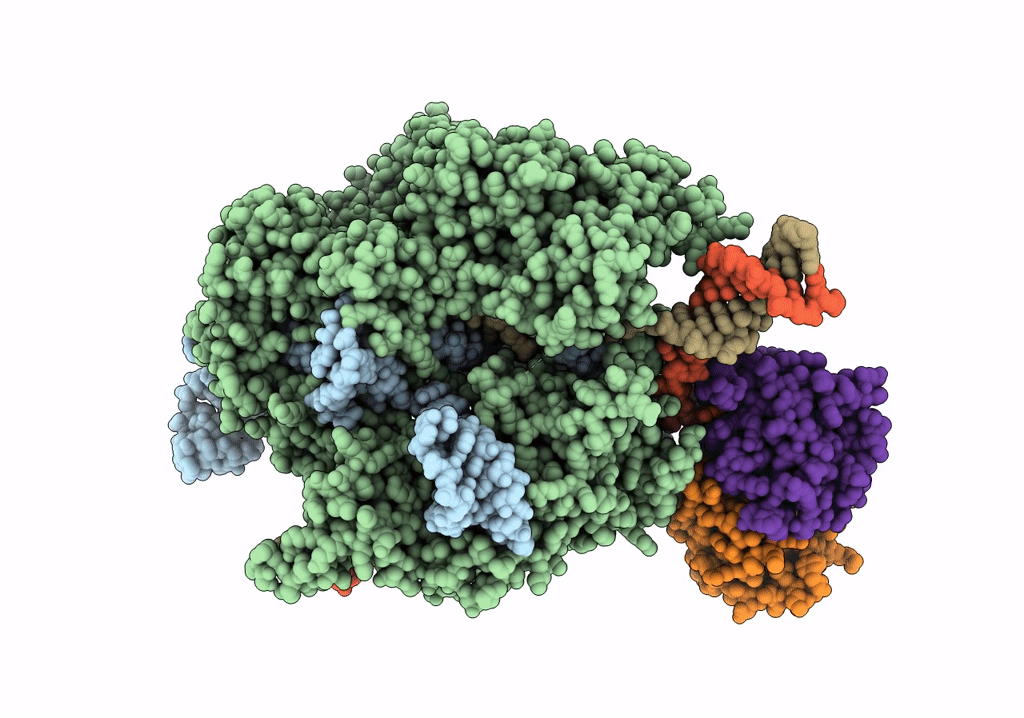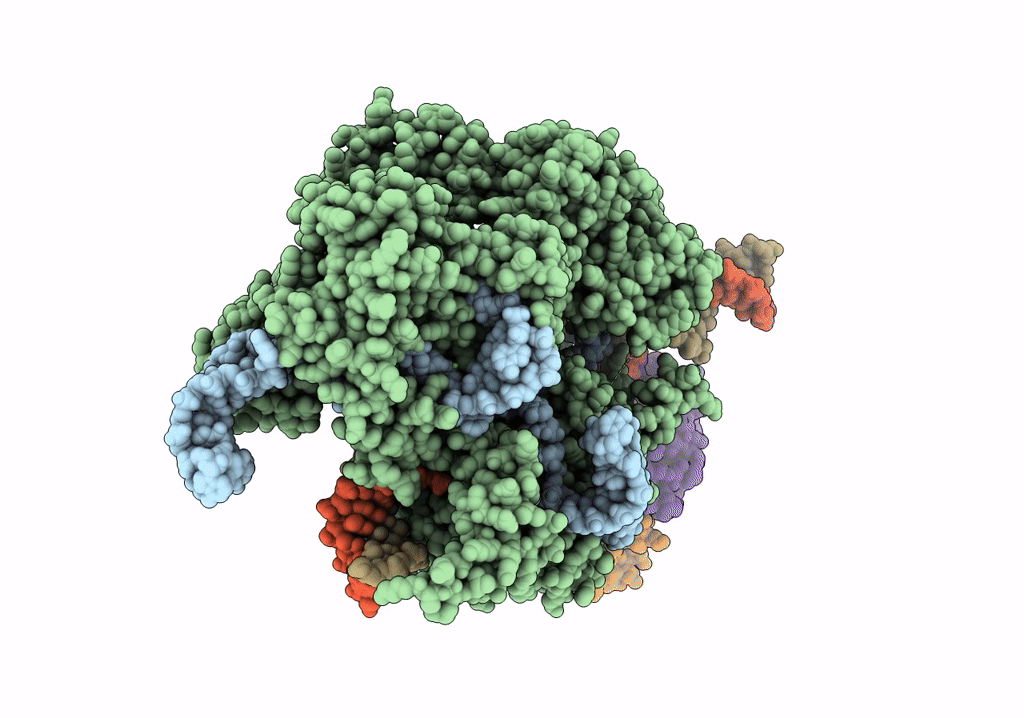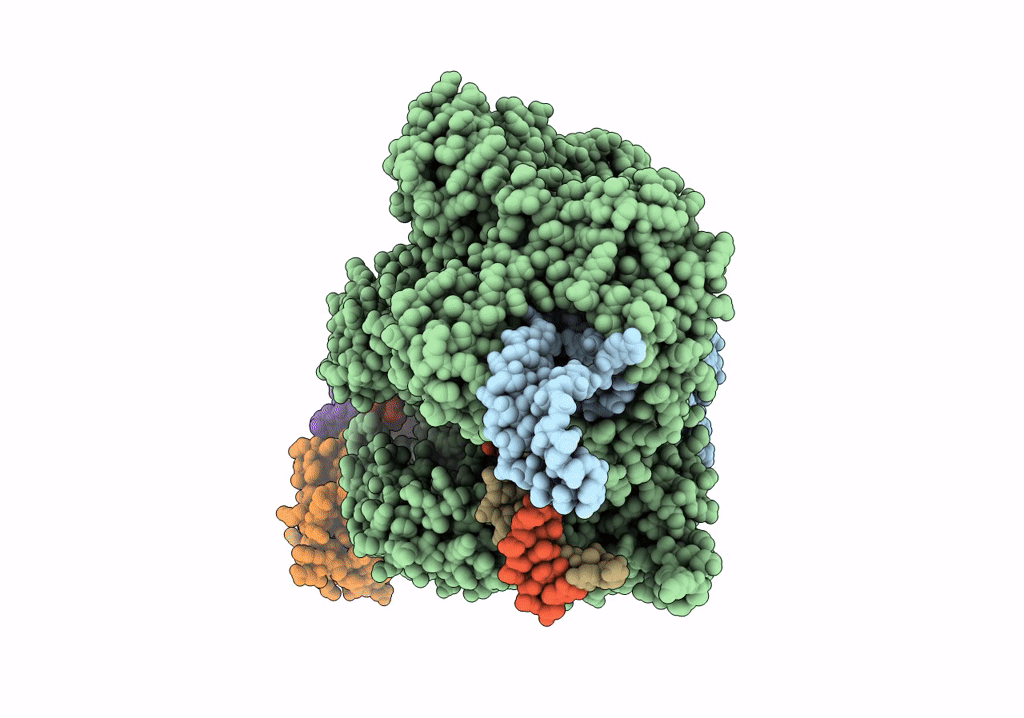
Structure of the SpCas9 DNA adenine base editor - ABE8e
Biological Source:
Source Organism(s):
Streptococcus pyogenes (Taxon ID: 1314)
Escherichia coli (Taxon ID: 562)
synthetic construct (Taxon ID: 32630)
Escherichia coli (Taxon ID: 562)
synthetic construct (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
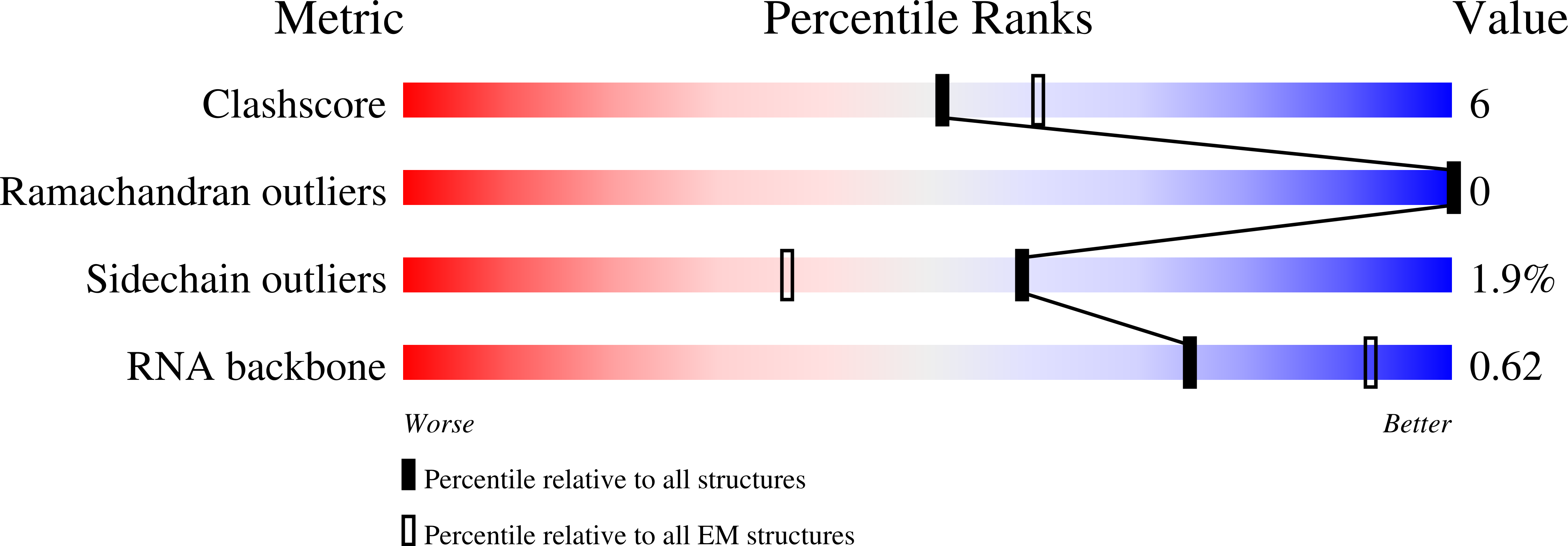
Resolution:
3.20 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE