Deposition Date
2020-01-16
Release Date
2021-04-21
Last Version Date
2023-11-15
Entry Detail
PDB ID:
6VJO
Keywords:
Title:
Human parainfluenza virus type 3 fusion glycoprotein N-terminal heptad repeat domain+alpha/beta-VI
Biological Source:
Source Organism(s):
Human respirovirus 3 (Taxon ID: 11216)
Method Details:
Experimental Method:
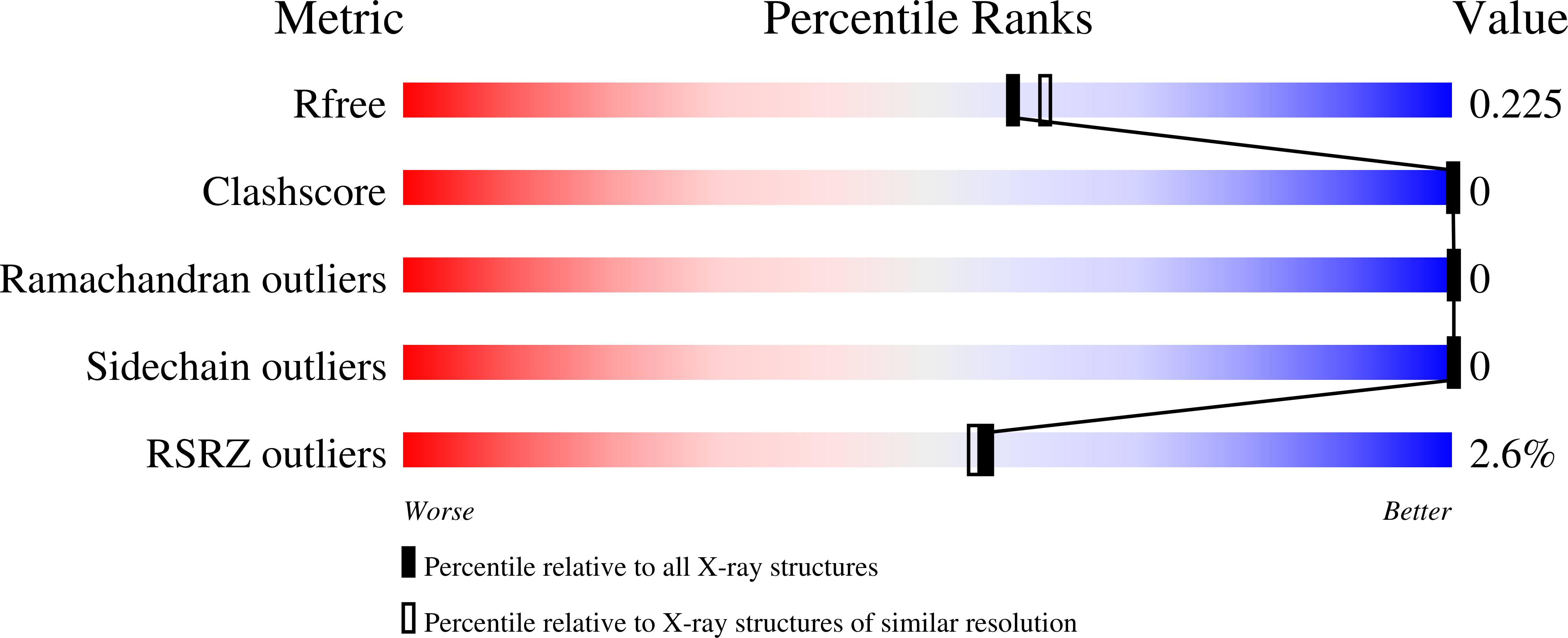
Resolution:
2.00 Å
R-Value Free:
0.22
R-Value Work:
0.19
R-Value Observed:
0.20
Space Group:
H 3