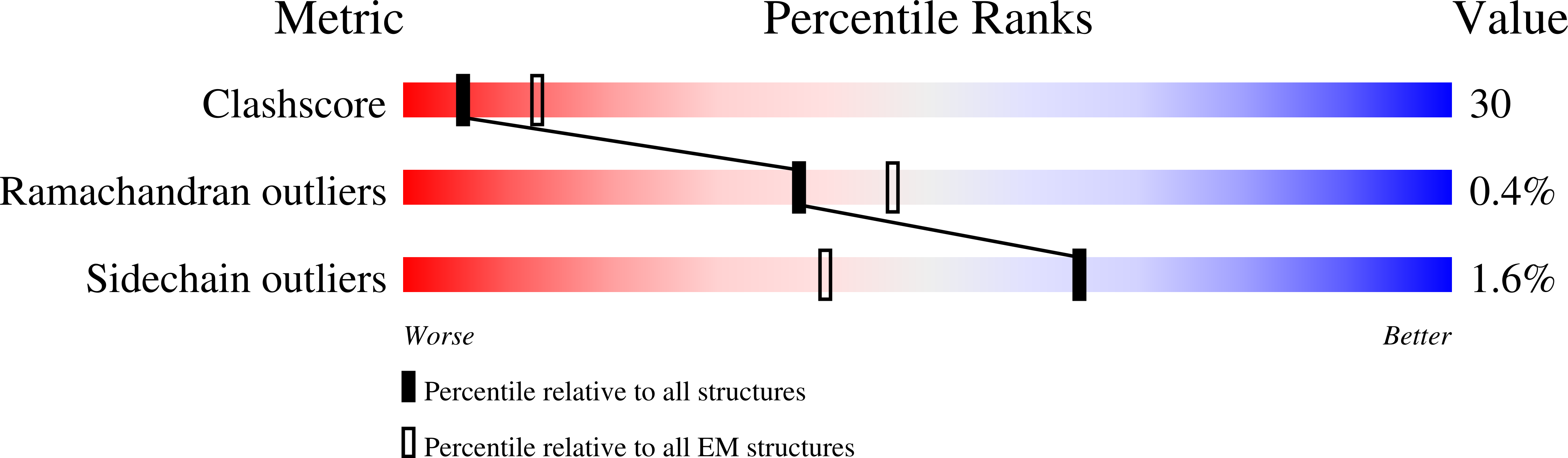
Deposition Date
2019-11-14
Release Date
2020-01-01
Last Version Date
2024-10-16
Entry Detail
Biological Source:
Source Organism:
Rattus norvegicus (Taxon ID: 10116)
Aequorea victoria (Taxon ID: 6100)
Aequorea victoria (Taxon ID: 6100)
Host Organism:
Method Details:
Experimental Method:
Resolution:
3.50 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE