Deposition Date
2019-11-13
Release Date
2020-04-22
Last Version Date
2024-11-20
Entry Detail
PDB ID:
6UYG
Keywords:
Title:
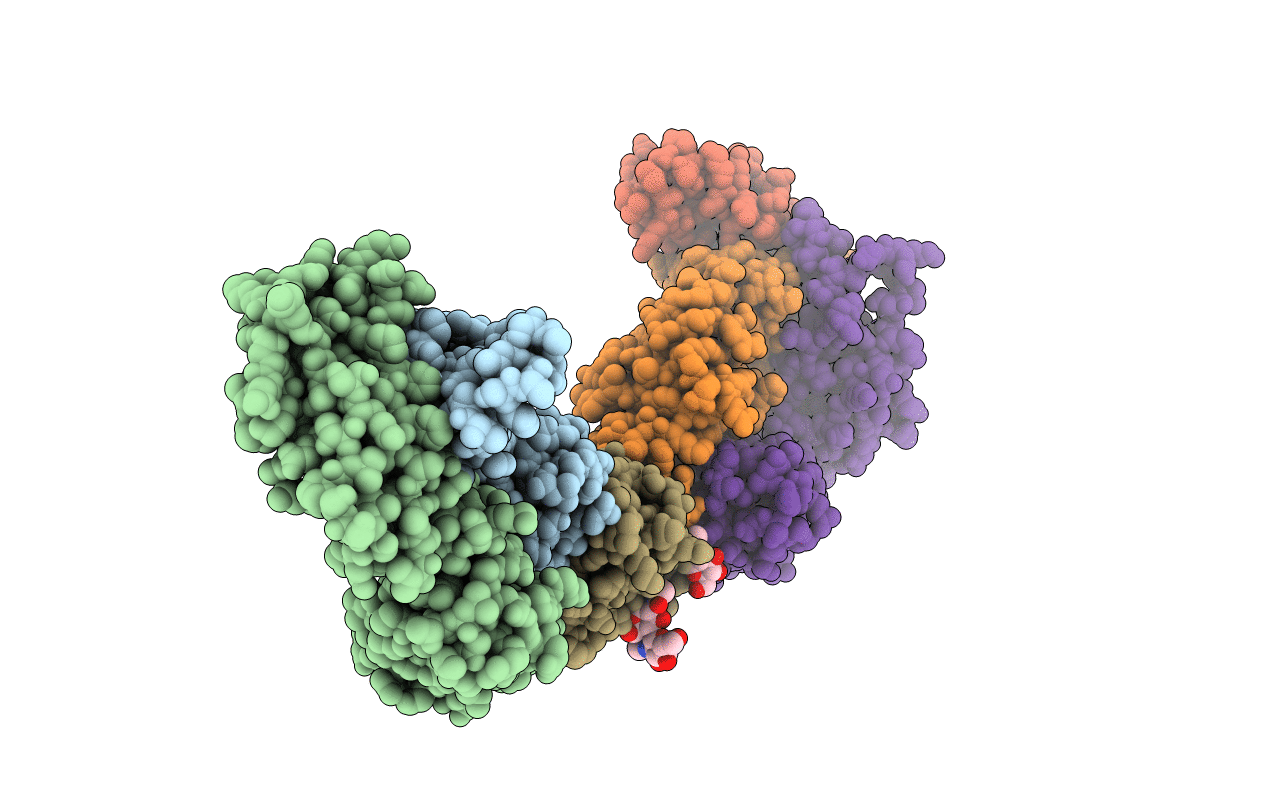
Structure of Hepatitis C Virus Envelope Glycoprotein E2c3 core from genotype 6a bound to broadly neutralizing antibody AR3A and non neutralizing antibody E1
Biological Source:
Source Organism:
Recombinant Hepatitis C virus HK6a/JFH-1 (Taxon ID: 595609)
Homo sapiens (Taxon ID: 9606)
Streptococcus sp. (Taxon ID: 1306)
Homo sapiens (Taxon ID: 9606)
Streptococcus sp. (Taxon ID: 1306)
Host Organism:
Method Details:
Experimental Method:
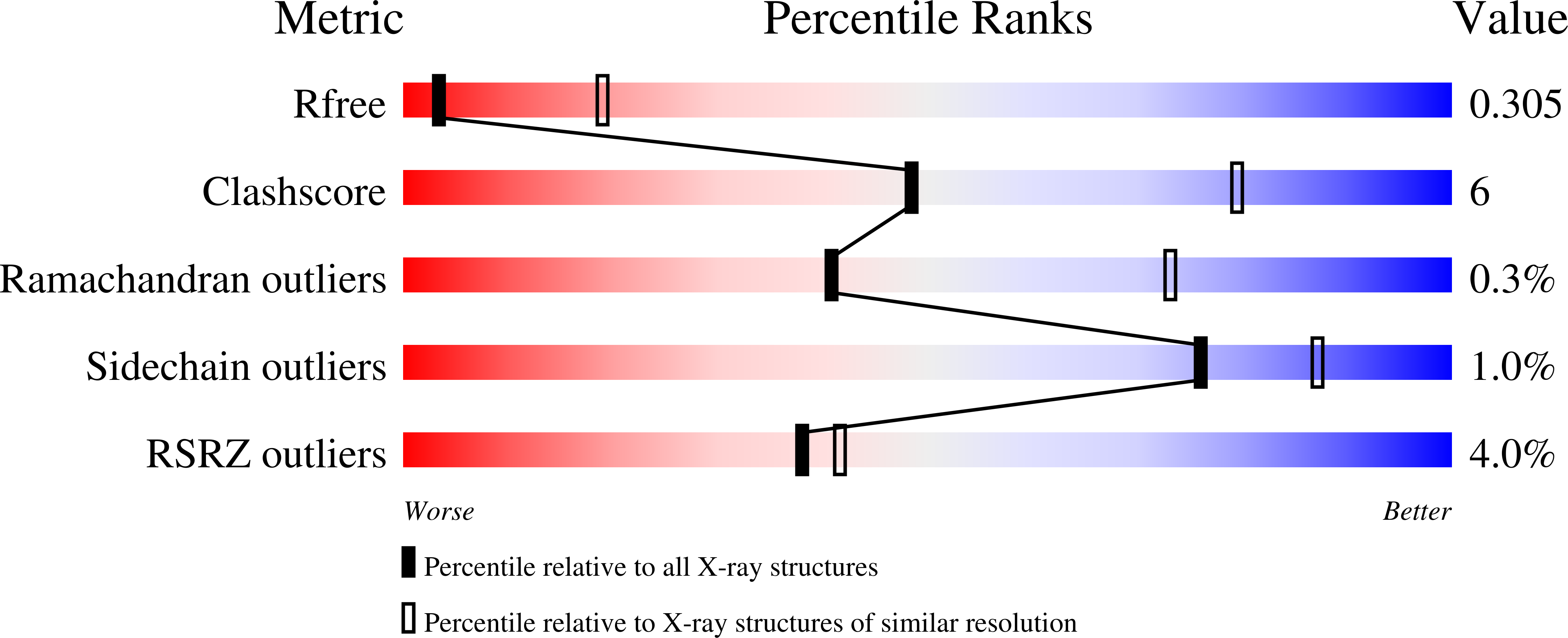
Resolution:
3.38 Å
R-Value Free:
0.30
R-Value Work:
0.25
R-Value Observed:
0.25
Space Group:
P 63 2 2