Deposition Date
2019-10-14
Release Date
2019-12-04
Last Version Date
2023-10-11
Entry Detail
PDB ID:
6UO7
Keywords:
Title:
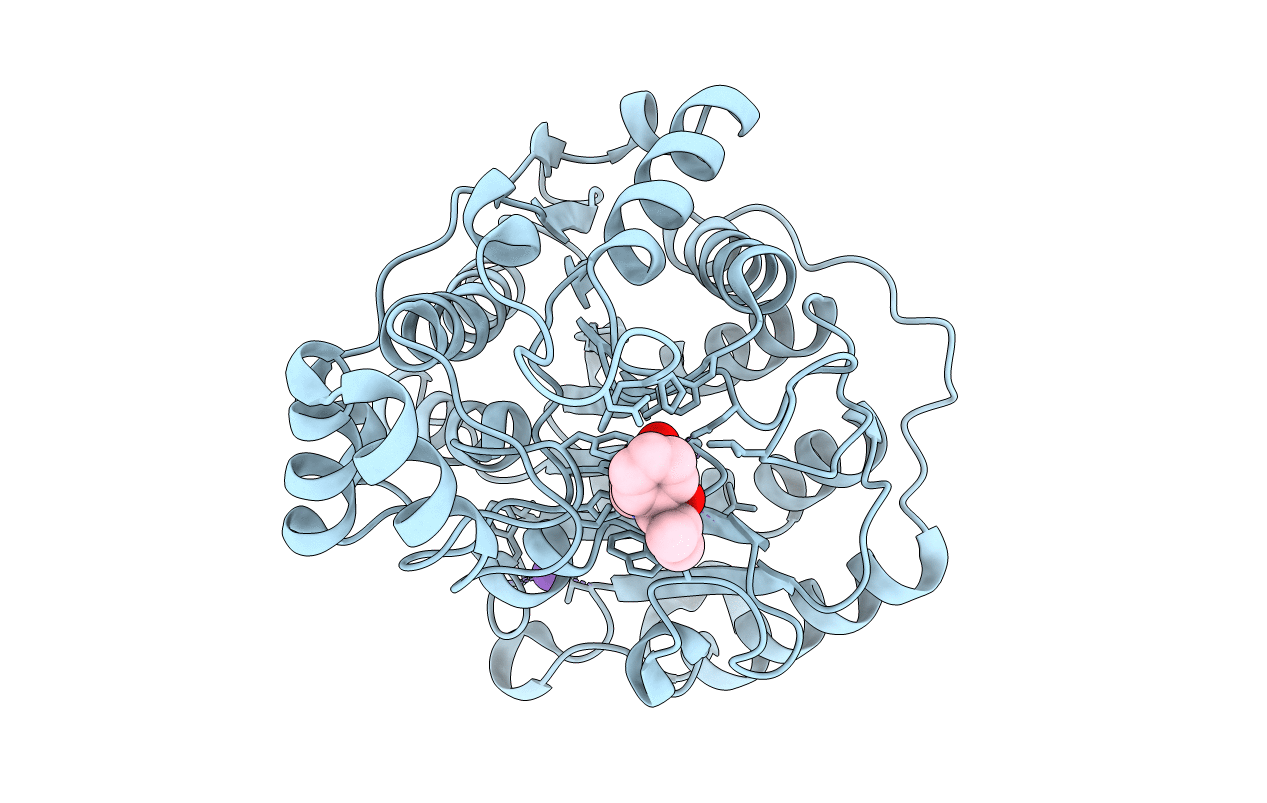
Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 1 (CD1) K330L mutant complexed with AR-42
Biological Source:
Source Organism:
Danio rerio (Taxon ID: 7955)
Host Organism:
Method Details:
Experimental Method:
Resolution:
1.40 Å
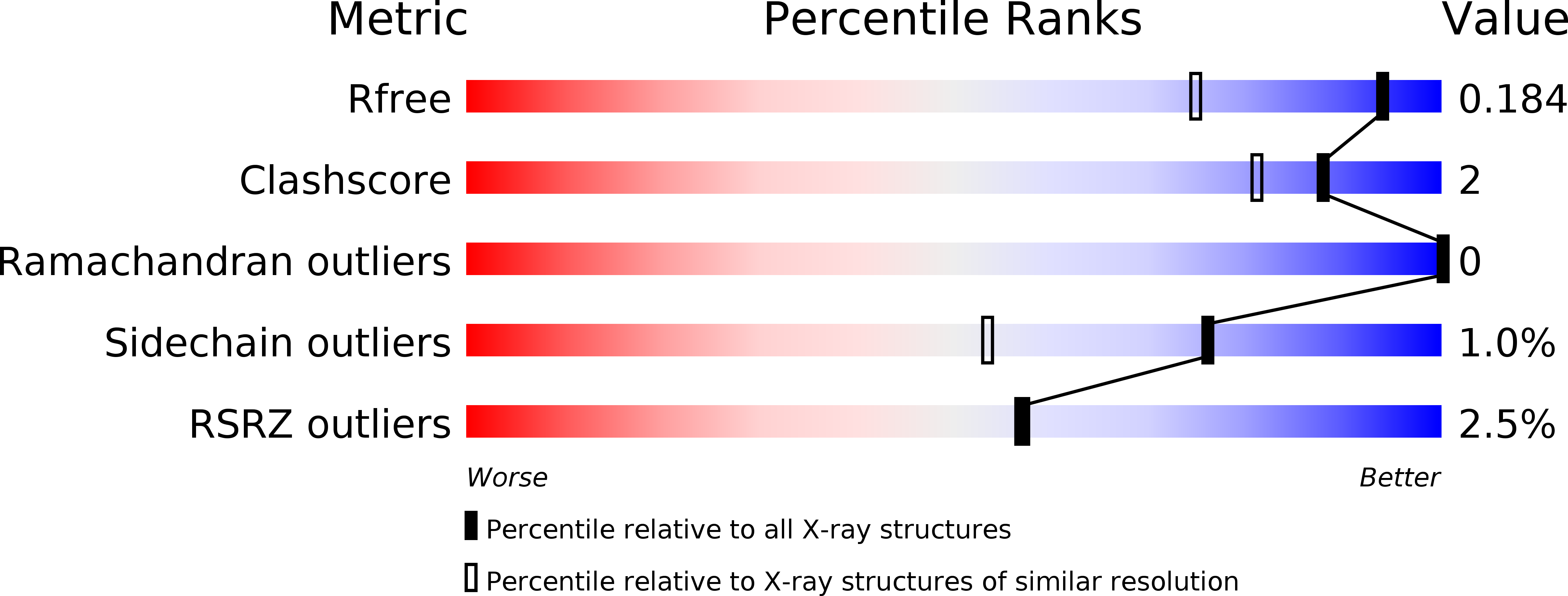
R-Value Free:
0.18
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
P 21 21 21